Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004962A_C01 KMC004962A_c01
(639 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
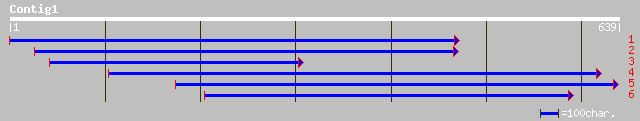
dbj|BAB55655.1| fructose-6-phosphate 2-kinase/fructose-2,6-bisph... 178 6e-44
pir||T08994 6-phosphofructo-2-kinase (EC 2.7.1.105) / fructose-2... 177 8e-44
gb|AAF04293.2|AF190739_1 fructose-6-phosphate 2-kinase/fructose-... 176 2e-43
ref|NP_172191.1| fructose-2,6-bisphosphatase, putative; protein ... 176 2e-43
gb|AAM91549.1| fructose-2,6-bisphosphatase, putative [Arabidopsi... 176 2e-43
>dbj|BAB55655.1| fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase
[Bruguiera gymnorrhiza]
Length = 745
Score = 178 bits (451), Expect = 6e-44
Identities = 88/90 (97%), Positives = 90/90 (99%)
Frame = -3
Query: 637 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 458
RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPL+E
Sbjct: 656 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLRE 715
Query: 457 IPHIEMPLHTIIEIQMGVTGVQEKRYKLMD 368
IPHIE+PLHTIIEIQMGVTGVQEKRYKLMD
Sbjct: 716 IPHIEVPLHTIIEIQMGVTGVQEKRYKLMD 745
>pir||T08994 6-phosphofructo-2-kinase (EC 2.7.1.105) / fructose-2,
6-bisphosphate 2-phosphatase (EC 3.1.3.46) - spinach
gi|3170230|gb|AAC18055.1| fructose-6-phosphate 2-kinase
/fructose-2,6-bisphosphatase [Spinacia oleracea]
Length = 750
Score = 177 bits (450), Expect = 8e-44
Identities = 88/90 (97%), Positives = 90/90 (99%)
Frame = -3
Query: 637 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 458
R+KDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE
Sbjct: 661 REKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 720
Query: 457 IPHIEMPLHTIIEIQMGVTGVQEKRYKLMD 368
IPHIE+PLHTIIEIQMGVTGVQEKRYKLMD
Sbjct: 721 IPHIEVPLHTIIEIQMGVTGVQEKRYKLMD 750
>gb|AAF04293.2|AF190739_1 fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase
[Arabidopsis thaliana]
Length = 744
Score = 176 bits (446), Expect = 2e-43
Identities = 88/90 (97%), Positives = 89/90 (98%)
Frame = -3
Query: 637 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 458
RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE
Sbjct: 655 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 714
Query: 457 IPHIEMPLHTIIEIQMGVTGVQEKRYKLMD 368
IP IEMPLHTIIEIQMGV+GVQEKRYKLMD
Sbjct: 715 IPQIEMPLHTIIEIQMGVSGVQEKRYKLMD 744
>ref|NP_172191.1| fructose-2,6-bisphosphatase, putative; protein id: At1g07110.1,
supported by cDNA: gi_13096097, supported by cDNA:
gi_8117171 [Arabidopsis thaliana]
gi|8117172|dbj|BAA96353.1|
fructose-6-phosphate,2-kinase/fructose-2,
6-bisphosphatase [Arabidopsis thaliana]
Length = 744
Score = 176 bits (446), Expect = 2e-43
Identities = 88/90 (97%), Positives = 89/90 (98%)
Frame = -3
Query: 637 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 458
RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE
Sbjct: 655 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 714
Query: 457 IPHIEMPLHTIIEIQMGVTGVQEKRYKLMD 368
IP IEMPLHTIIEIQMGV+GVQEKRYKLMD
Sbjct: 715 IPQIEMPLHTIIEIQMGVSGVQEKRYKLMD 744
>gb|AAM91549.1| fructose-2,6-bisphosphatase, putative [Arabidopsis thaliana]
Length = 355
Score = 176 bits (446), Expect = 2e-43
Identities = 88/90 (97%), Positives = 89/90 (98%)
Frame = -3
Query: 637 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 458
RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE
Sbjct: 266 RKKDKLRYRYPRGESYLDVIQRLEPVIIELERQRAPVVVISHQAVLRALYAYFADRPLKE 325
Query: 457 IPHIEMPLHTIIEIQMGVTGVQEKRYKLMD 368
IP IEMPLHTIIEIQMGV+GVQEKRYKLMD
Sbjct: 326 IPQIEMPLHTIIEIQMGVSGVQEKRYKLMD 355
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 518,922,288
Number of Sequences: 1393205
Number of extensions: 10589297
Number of successful extensions: 19793
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 19257
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19708
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26723359358
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)