Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004954A_C01 KMC004954A_c01
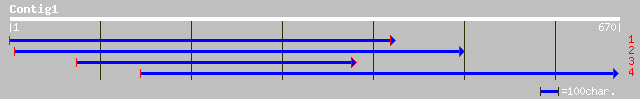
(670 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK28804.1|AF310959_1 resistance-like protein P1-B [Linum usi... 71 1e-11
gb|AAK28805.1|AF310960_1 resistance-like protein P2-A [Linum usi... 71 1e-11
gb|AAK28808.1|AF310961_1 resistance-like protein P3-A [Linum usi... 71 1e-11
gb|AAK28803.1|AF310958_1 resistance-like protein P1-A [Linum usi... 68 1e-10
gb|AAK28811.1|AF310966_1 resistance-like protein P-B [Linum usit... 67 2e-10
>gb|AAK28804.1|AF310959_1 resistance-like protein P1-B [Linum usitatissimum]
Length = 1196
Score = 71.2 bits (173), Expect = 1e-11
Identities = 43/107 (40%), Positives = 66/107 (61%), Gaps = 1/107 (0%)
Frame = -3
Query: 644 LNLSDTGIKALPSSFGSQRKLEILDLEQGR-IESLPSCIVNLTRLRYLNLSDCEKLQTLP 468
L LS TGIK+LPSS R+L +DL + +ES+P+ I NL+ L ++S C+ + +LP
Sbjct: 920 LRLSKTGIKSLPSSIHELRQLYSIDLRNCKSLESIPNSIHNLSSLVTFSMSGCKIIISLP 979
Query: 467 ELPPSVETLLTRGCSSLKTVLFPSTATEQSQENKKRVEFFQCESLDE 327
ELPP+++TL GC SL+ + PS + N R+ F +C +D+
Sbjct: 980 ELPPNLKTLNVSGCKSLQAL--PSNTCKLLYLN--RIYFEECPQVDQ 1022
>gb|AAK28805.1|AF310960_1 resistance-like protein P2-A [Linum usitatissimum]
Length = 1196
Score = 71.2 bits (173), Expect = 1e-11
Identities = 44/111 (39%), Positives = 68/111 (60%), Gaps = 1/111 (0%)
Frame = -3
Query: 656 NMIELNLSDTGIKALPSSFGSQRKLEILDLEQGR-IESLPSCIVNLTRLRYLNLSDCEKL 480
++I L L +TGIK+LPSS R+L +DL + +ES+P+ I L++L L++S CE +
Sbjct: 917 SLISLCLVETGIKSLPSSIQELRQLFSIDLRDCKSLESIPNSIHKLSKLVTLSMSGCEII 976
Query: 479 QTLPELPPSVETLLTRGCSSLKTVLFPSTATEQSQENKKRVEFFQCESLDE 327
+LPELPP+++TL GC SL+ + PS + N + F C LD+
Sbjct: 977 ISLPELPPNLKTLNVSGCKSLQAL--PSNTCKLLYLN--TIHFDGCPQLDQ 1023
>gb|AAK28808.1|AF310961_1 resistance-like protein P3-A [Linum usitatissimum]
Length = 1110
Score = 71.2 bits (173), Expect = 1e-11
Identities = 44/111 (39%), Positives = 68/111 (60%), Gaps = 1/111 (0%)
Frame = -3
Query: 656 NMIELNLSDTGIKALPSSFGSQRKLEILDLEQGR-IESLPSCIVNLTRLRYLNLSDCEKL 480
++I L L +TGIK+LPSS R+L +DL + +ES+P+ I L++L L++S CE +
Sbjct: 915 SLISLCLVETGIKSLPSSIQELRQLFSIDLRDCKSLESIPNSIHKLSKLVTLSMSGCEII 974
Query: 479 QTLPELPPSVETLLTRGCSSLKTVLFPSTATEQSQENKKRVEFFQCESLDE 327
+LPELPP+++TL GC SL+ + PS + N + F C LD+
Sbjct: 975 ISLPELPPNLKTLNVSGCKSLQAL--PSNTCKLLYLN--TIHFDGCPQLDQ 1021
>gb|AAK28803.1|AF310958_1 resistance-like protein P1-A [Linum usitatissimum]
Length = 1200
Score = 67.8 bits (164), Expect = 1e-10
Identities = 42/109 (38%), Positives = 65/109 (59%), Gaps = 1/109 (0%)
Frame = -3
Query: 644 LNLSDTGIKALPSSFGSQRKLEILDLEQGR-IESLPSCIVNLTRLRYLNLSDCEKLQTLP 468
L L +TGIK+LPSS R+L +DL + +ES+P+ I L++L ++S CE + +LP
Sbjct: 923 LYLVETGIKSLPSSIQELRQLYSIDLRDCKSLESIPNSIHKLSKLVTFSMSGCESIPSLP 982
Query: 467 ELPPSVETLLTRGCSSLKTVLFPSTATEQSQENKKRVEFFQCESLDERS 321
ELPP+++ L C SL+ + PS + N R+ F +C LD+ S
Sbjct: 983 ELPPNLKELDVSRCKSLQAL--PSNTCKLWYLN--RIYFEECPQLDQTS 1027
>gb|AAK28811.1|AF310966_1 resistance-like protein P-B [Linum usitatissimum]
Length = 1211
Score = 67.4 bits (163), Expect = 2e-10
Identities = 40/107 (37%), Positives = 64/107 (59%), Gaps = 1/107 (0%)
Frame = -3
Query: 644 LNLSDTGIKALPSSFGSQRKLEILDLEQGR-IESLPSCIVNLTRLRYLNLSDCEKLQTLP 468
L L +TGIK+LPSS R+L + L + +ES+P+ I L++L ++S CE + +LP
Sbjct: 935 LRLVETGIKSLPSSIHELRQLHSICLRDCKSLESIPNSIHKLSKLGTFSMSGCESIPSLP 994
Query: 467 ELPPSVETLLTRGCSSLKTVLFPSTATEQSQENKKRVEFFQCESLDE 327
ELPP+++ L R C SL+ + PS + N R+ F +C +D+
Sbjct: 995 ELPPNLKELEVRDCKSLQAL--PSNTCKLLYLN--RIYFEECPQVDQ 1037
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,783,382
Number of Sequences: 1393205
Number of extensions: 11059752
Number of successful extensions: 35719
Number of sequences better than 10.0: 939
Number of HSP's better than 10.0 without gapping: 32185
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35289
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)