Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004943A_C01 KMC004943A_c01
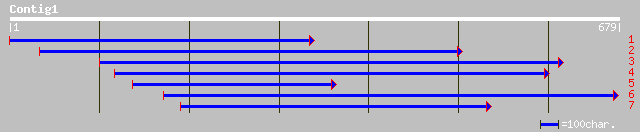
(675 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_174112.1| unknown protein; protein id: At1g27910.1, suppo... 109 3e-23
ref|NP_173843.1| hypothetical protein; protein id: At1g24330.1 [... 103 2e-21
ref|NP_176920.1| hypothetical protein; protein id: At1g67530.1 [... 97 2e-19
dbj|BAB89956.1| P0035F12.13 [Oryza sativa (japonica cultivar-gro... 63 3e-09
emb|CAD41127.1| OSJNBa0084K20.7 [Oryza sativa (japonica cultivar... 56 4e-07
>ref|NP_174112.1| unknown protein; protein id: At1g27910.1, supported by cDNA:
gi_20453194 [Arabidopsis thaliana]
gi|25403044|pir||D86404 unknown protein [imported] -
Arabidopsis thaliana
gi|12322984|gb|AAG51474.1|AC069471_5 unknown protein
[Arabidopsis thaliana] gi|20453195|gb|AAM19837.1|
At1g27910/F13K9_2 [Arabidopsis thaliana]
gi|27363436|gb|AAO11637.1| At1g27910/F13K9_2
[Arabidopsis thaliana]
Length = 768
Score = 109 bits (273), Expect = 3e-23
Identities = 60/98 (61%), Positives = 68/98 (69%), Gaps = 6/98 (6%)
Frame = -1
Query: 675 VLQEGVIPALVSISVNGTPRGREKAQKLLMLFREQRQRDHS----PVETHQSPPETG--D 514
VLQEGVIP+LVSISVNGT RGRE+AQKLL LFRE RQRD + P T + PE G
Sbjct: 671 VLQEGVIPSLVSISVNGTQRGRERAQKLLTLFRELRQRDQTHLTEPQHTEVTSPEDGFSV 730
Query: 513 LSVPPAEMKPLCKSMSRRKTMKGFGFFWKTKSYSVYQC 400
S E KP CKS SR+K + F F WK+KS+SVYQC
Sbjct: 731 ASAAVTETKPQCKSASRKKMGRAFSFLWKSKSFSVYQC 768
>ref|NP_173843.1| hypothetical protein; protein id: At1g24330.1 [Arabidopsis
thaliana] gi|7486373|pir||T00664 hypothetical protein
F3I6.27 - Arabidopsis thaliana gi|2829887|gb|AAC00595.1|
Hypothetical protein [Arabidopsis thaliana]
Length = 709
Score = 103 bits (257), Expect = 2e-21
Identities = 57/99 (57%), Positives = 72/99 (72%), Gaps = 9/99 (9%)
Frame = -1
Query: 675 VLQEGVIPALVSISVNGTPRGREKAQKLLMLFREQRQRDHSPVETHQSPPET-------- 520
VLQEGVIP+LVSISVNG+PRGR+K+QKLLMLFREQR RD ++P +T
Sbjct: 611 VLQEGVIPSLVSISVNGSPRGRDKSQKLLMLFREQRHRDQPSPNKEEAPRKTVSAPMAIP 670
Query: 519 GDLSVPPAEMKPLCKSMSRRKTM-KGFGFFWKTKSYSVY 406
+S P +E+KPL KS+SRRKTM + F F WK KS+S++
Sbjct: 671 APVSAPESEVKPLTKSISRRKTMTRPFSFLWK-KSHSIH 708
>ref|NP_176920.1| hypothetical protein; protein id: At1g67530.1 [Arabidopsis
thaliana] gi|25404676|pir||G96698 hypothetical protein
F12B7.8 [imported] - Arabidopsis thaliana
gi|12324681|gb|AAG52304.1|AC011020_11 hypothetical
protein [Arabidopsis thaliana]
gi|26449494|dbj|BAC41873.1| unknown protein [Arabidopsis
thaliana]
Length = 782
Score = 97.4 bits (241), Expect = 2e-19
Identities = 59/108 (54%), Positives = 67/108 (61%), Gaps = 19/108 (17%)
Frame = -1
Query: 675 VLQEGVIPALVSISVNGTPRGREKAQKLLMLFREQRQRDHSPVETHQSPPE--------- 523
VLQEGVIP+LVSISVNGTPRGREK+QKLLMLFRE+RQ+ P PP+
Sbjct: 674 VLQEGVIPSLVSISVNGTPRGREKSQKLLMLFREERQQRDQPSSNRDEPPQKEPARKSLS 733
Query: 522 ---------TGDLSVPPAEMKPLCKSMSRRKTM-KGFGFFWKTKSYSV 409
SV E + L KSMSRRK+M + F FFWK KSYSV
Sbjct: 734 APLSVHGSTPASASVQDYEPRVLSKSMSRRKSMARPFSFFWK-KSYSV 780
>dbj|BAB89956.1| P0035F12.13 [Oryza sativa (japonica cultivar-group)]
gi|20161603|dbj|BAB90523.1| B1065G12.5 [Oryza sativa
(japonica cultivar-group)]
Length = 826
Score = 63.2 bits (152), Expect = 3e-09
Identities = 30/39 (76%), Positives = 35/39 (88%)
Frame = -1
Query: 675 VLQEGVIPALVSISVNGTPRGREKAQKLLMLFREQRQRD 559
VLQEGV+P+LVSIS GT +G+EK+QKLL LFREQRQRD
Sbjct: 713 VLQEGVVPSLVSISATGTGKGKEKSQKLLKLFREQRQRD 751
>emb|CAD41127.1| OSJNBa0084K20.7 [Oryza sativa (japonica cultivar-group)]
Length = 861
Score = 56.2 bits (134), Expect = 4e-07
Identities = 26/39 (66%), Positives = 35/39 (89%)
Frame = -1
Query: 675 VLQEGVIPALVSISVNGTPRGREKAQKLLMLFREQRQRD 559
VLQEGVIPALVS++ NGT + ++KAQ+LL+LFR +RQR+
Sbjct: 756 VLQEGVIPALVSLTANGTGKTKDKAQRLLLLFRGKRQRE 794
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 615,081,085
Number of Sequences: 1393205
Number of extensions: 14153082
Number of successful extensions: 44182
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 41564
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44073
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29704274460
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)