Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004941A_C01 KMC004941A_c01
(570 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
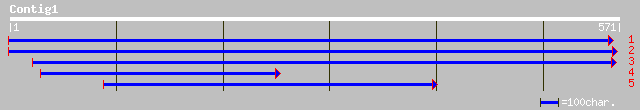
emb|CAD56218.1| hypothetical protein [Cicer arietinum] 138 6e-32
ref|NP_186824.1| hypothetical protein; protein id: At3g01750.1, ... 66 3e-10
gb|AAK76397.1| Rev-Erb beta [Oncorhynchus mykiss] 39 0.060
ref|NP_594130.1| putative glucoamylase I (alpha-1,4-glucan gluco... 34 1.5
ref|NP_595617.1| hnf-3/fork-head transcription factor homolog [S... 32 4.3
>emb|CAD56218.1| hypothetical protein [Cicer arietinum]
Length = 322
Score = 138 bits (347), Expect = 6e-32
Identities = 82/126 (65%), Positives = 91/126 (72%), Gaps = 6/126 (4%)
Frame = -3
Query: 568 LSVRSHQSSPNAKKRFASLVHGVMQSLPQAKVSVSGRSRSSSFSKSSISSPSSIDKPKGI 389
LSVRSHQSSPNAKKRFA GRSRSSSFSKSSISSP SIDK KGI
Sbjct: 214 LSVRSHQSSPNAKKRFAC-----------------GRSRSSSFSKSSISSPRSIDKQKGI 256
Query: 388 C-IDNDL-AGPSCSSELPE----DESPHLGKRTSFSKKLRGHYLCFGAPGLNVKSSVHKA 227
I+ND+ +GPSCSS+L + DE P L KR S S+KL+GHY CFGAPGLNVK+SVHKA
Sbjct: 257 IYINNDIVSGPSCSSQLRDRGGDDELPKLVKRNSVSRKLKGHYFCFGAPGLNVKNSVHKA 316
Query: 226 NVVAVA 209
+ VAVA
Sbjct: 317 SFVAVA 322
>ref|NP_186824.1| hypothetical protein; protein id: At3g01750.1, supported by cDNA:
gi_17529251, supported by cDNA: gi_20465972 [Arabidopsis
thaliana] gi|6016732|gb|AAF01558.1|AC009325_28
hypothetical protein [Arabidopsis thaliana]
gi|6091719|gb|AAF03431.1|AC010797_7 hypothetical protein
[Arabidopsis thaliana] gi|17529252|gb|AAL38853.1|
unknown protein [Arabidopsis thaliana]
gi|20465973|gb|AAM20172.1| unknown protein [Arabidopsis
thaliana]
Length = 664
Score = 65.9 bits (159), Expect = 3e-10
Identities = 47/117 (40%), Positives = 60/117 (51%), Gaps = 8/117 (6%)
Frame = -3
Query: 568 LSVRSHQSSPNA-KKRFASLVHGVMQSLPQAKVSVSGRSRSSSFSKSSISSPSSID---- 404
L+VRS+QSSP A KKRF S+ RSRSSSFSK SI S S+
Sbjct: 559 LAVRSNQSSPRAKKKRFGSV-----------------RSRSSSFSKISIHSSSTSSTSSM 601
Query: 403 ---KPKGICIDNDLAGPSCSSELPEDESPHLGKRTSFSKKLRGHYLCFGAPGLNVKS 242
K KG+ ID +AGPS ++ K+ S +L+ HY CFG L+VK+
Sbjct: 602 VDLKQKGVLIDTSIAGPSGLKRSEPVKNKVSEKQGSVRSRLKSHYFCFGTSALSVKT 658
>gb|AAK76397.1| Rev-Erb beta [Oncorhynchus mykiss]
Length = 461
Score = 38.5 bits (88), Expect = 0.060
Identities = 27/77 (35%), Positives = 37/77 (47%), Gaps = 6/77 (7%)
Frame = -3
Query: 538 NAKKRFASLVHGV------MQSLPQAKVSVSGRSRSSSFSKSSISSPSSIDKPKGICIDN 377
N + S++HG M+ LP S S S SSS S SS S+PSS + +
Sbjct: 75 NNNSQHHSMLHGTQSPPCPMEGLPNDGSSSSSASSSSSISSSSGSNPSSPCSAQDSMETS 134
Query: 376 DLAGPSCSSELPEDESP 326
+ SCSS+ +DE P
Sbjct: 135 SSSASSCSSDSGDDEVP 151
>ref|NP_594130.1| putative glucoamylase I (alpha-1,4-glucan glucosidase),
extracellular starch-degrading enzyme; by similarity to
S. cerevisiae STA1; contains chitinase family signature
[Schizosaccharomyces pombe] gi|13624759|emb|CAC36921.1|
putative glucoamylase I (alpha-1,4-glucan glucosidase),
extracellular starch-degrading enzyme; by similarity to
S. cerevisiae STA1; contains chitinase family signature
[Schizosaccharomyces pombe]
Length = 1236
Score = 33.9 bits (76), Expect = 1.5
Identities = 19/50 (38%), Positives = 27/50 (54%)
Frame = -3
Query: 556 SHQSSPNAKKRFASLVHGVMQSLPQAKVSVSGRSRSSSFSKSSISSPSSI 407
S S P+ +S++ S +S+S S SS+FS +S SSPSSI
Sbjct: 565 SSSSIPSTFSSVSSILSSSTSSPSSTSLSISSSSTSSTFSSASTSSPSSI 614
>ref|NP_595617.1| hnf-3/fork-head transcription factor homolog [Schizosaccharomyces
pombe] gi|13629413|sp|O13606|MEI4_SCHPO Meiosis-specific
transcription factor mei4 gi|11359057|pir||T43358
hnf-3/forkhead transcription factor - fission yeast
(Schizosaccharomyces pombe) gi|2257493|dbj|BAA21390.1|
miosis-specific transcription factor
[Schizosaccharomyces pombe] gi|3036967|dbj|BAA25402.1|
forkhead/HNF3 homologue [Schizosaccharomyces pombe]
gi|13872529|emb|CAC37501.1| fork head transcription
factor homolog; meiosis specific transcription factor
[Schizosaccharomyces pombe]
Length = 517
Score = 32.3 bits (72), Expect = 4.3
Identities = 22/67 (32%), Positives = 34/67 (49%)
Frame = -3
Query: 559 RSHQSSPNAKKRFASLVHGVMQSLPQAKVSVSGRSRSSSFSKSSISSPSSIDKPKGICID 380
RSH + N+KKR +S H + + RSR +SF+ S+ +S SS + +
Sbjct: 172 RSHSTDSNSKKRPSSKCHEIKPLTTREIPLARKRSRLNSFNSSTSTSGSSSNVAAE--VS 229
Query: 379 NDLAGPS 359
ND + PS
Sbjct: 230 NDASQPS 236
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,904,124
Number of Sequences: 1393205
Number of extensions: 7983472
Number of successful extensions: 21338
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 20503
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21244
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)