Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004930A_C01 KMC004930A_c01
(510 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
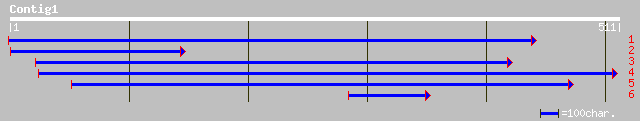
ref|NP_568314.1| HSP100/ClpB, putative; protein id: At5g15450.1 ... 173 9e-43
gb|AAN17424.1| HSP100/ClpB, putative [Arabidopsis thaliana] gi|2... 173 9e-43
gb|AAF91178.1|AF203700_1 ClpB [Phaseolus lunatus] 109 2e-23
pir||G84644 probable ATP-dependent CLPB protein [imported] - Ara... 108 3e-23
ref|NP_565586.1| HSP100/ClpB, putative; protein id: At2g25140.1,... 108 3e-23
>ref|NP_568314.1| HSP100/ClpB, putative; protein id: At5g15450.1 [Arabidopsis thaliana]
gi|11265210|pir||T51523 clpB heat shock protein-like -
Arabidopsis thaliana gi|9755800|emb|CAC01744.1| clpB heat
shock protein-like [Arabidopsis thaliana]
Length = 968
Score = 173 bits (439), Expect = 9e-43
Identities = 86/114 (75%), Positives = 102/114 (89%)
Frame = -3
Query: 508 RDQISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVEN 329
R+QI+ IVRLQL RVQKRIADRKMKI +T+AA+ LLG+LGYDPNYGARPVKRVIQQN+EN
Sbjct: 852 REQINRIVRLQLARVQKRIADRKMKINITDAAVDLLGSLGYDPNYGARPVKRVIQQNIEN 911
Query: 328 ELAKGILRGEFKDEDTILVDTEVTAFANGQLPQQKLVFRRVETDSKSTAKYSQE 167
ELAKGILRG+FK+ED IL+DTEVTAF+NGQLPQQKL F+++E++ TA QE
Sbjct: 912 ELAKGILRGDFKEEDGILIDTEVTAFSNGQLPQQKLTFKKIESE---TADAEQE 962
>gb|AAN17424.1| HSP100/ClpB, putative [Arabidopsis thaliana]
gi|27311927|gb|AAO00929.1| HSP100/ClpB, putative
[Arabidopsis thaliana]
Length = 173
Score = 173 bits (439), Expect = 9e-43
Identities = 86/114 (75%), Positives = 102/114 (89%)
Frame = -3
Query: 508 RDQISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVEN 329
R+QI+ IVRLQL RVQKRIADRKMKI +T+AA+ LLG+LGYDPNYGARPVKRVIQQN+EN
Sbjct: 57 REQINRIVRLQLARVQKRIADRKMKINITDAAVDLLGSLGYDPNYGARPVKRVIQQNIEN 116
Query: 328 ELAKGILRGEFKDEDTILVDTEVTAFANGQLPQQKLVFRRVETDSKSTAKYSQE 167
ELAKGILRG+FK+ED IL+DTEVTAF+NGQLPQQKL F+++E++ TA QE
Sbjct: 117 ELAKGILRGDFKEEDGILIDTEVTAFSNGQLPQQKLTFKKIESE---TADAEQE 167
>gb|AAF91178.1|AF203700_1 ClpB [Phaseolus lunatus]
Length = 977
Score = 109 bits (272), Expect = 2e-23
Identities = 51/82 (62%), Positives = 68/82 (82%)
Frame = -3
Query: 505 DQISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVENE 326
+QIS IV LQ+ERV+ R+ +K+ + TE A+K LG LG+DPN+GARPVKRVIQQ VENE
Sbjct: 867 EQISKIVELQMERVKNRLKQKKIDLHFTEEAVKHLGVLGFDPNFGARPVKRVIQQLVENE 926
Query: 325 LAKGILRGEFKDEDTILVDTEV 260
+A G+LRG+FK+ED+I+VD +V
Sbjct: 927 IAMGVLRGDFKEEDSIIVDADV 948
>pir||G84644 probable ATP-dependent CLPB protein [imported] - Arabidopsis thaliana
Length = 874
Score = 108 bits (271), Expect = 3e-23
Identities = 54/106 (50%), Positives = 78/106 (72%)
Frame = -3
Query: 505 DQISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVENE 326
++IS IV LQ+ RV+ + +K+K+Q T+ A+ LL LG+DPNYGARPVKRVIQQ VENE
Sbjct: 770 NEISKIVELQMRRVKNSLEQKKIKLQYTKEAVDLLAQLGFDPNYGARPVKRVIQQMVENE 829
Query: 325 LAKGILRGEFKDEDTILVDTEVTAFANGQLPQQKLVFRRVETDSKS 188
+A GIL+G+F +EDT+LVD + A N KLV +++E+++ +
Sbjct: 830 IAVGILKGDFAEEDTVLVDVDHLASDN------KLVIKKLESNASA 869
>ref|NP_565586.1| HSP100/ClpB, putative; protein id: At2g25140.1, supported by cDNA:
gi_17979453 [Arabidopsis thaliana]
gi|17979454|gb|AAL50064.1| At2g25140/F13D4.100
[Arabidopsis thaliana] gi|25090122|gb|AAN72234.1|
At2g25140/F13D4.100 [Arabidopsis thaliana]
Length = 964
Score = 108 bits (271), Expect = 3e-23
Identities = 54/106 (50%), Positives = 78/106 (72%)
Frame = -3
Query: 505 DQISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVENE 326
++IS IV LQ+ RV+ + +K+K+Q T+ A+ LL LG+DPNYGARPVKRVIQQ VENE
Sbjct: 860 NEISKIVELQMRRVKNSLEQKKIKLQYTKEAVDLLAQLGFDPNYGARPVKRVIQQMVENE 919
Query: 325 LAKGILRGEFKDEDTILVDTEVTAFANGQLPQQKLVFRRVETDSKS 188
+A GIL+G+F +EDT+LVD + A N KLV +++E+++ +
Sbjct: 920 IAVGILKGDFAEEDTVLVDVDHLASDN------KLVIKKLESNASA 959
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,661,728
Number of Sequences: 1393205
Number of extensions: 8726009
Number of successful extensions: 23842
Number of sequences better than 10.0: 392
Number of HSP's better than 10.0 without gapping: 23132
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23834
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15942513235
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)