Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004927A_C01 KMC004927A_c01
(847 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
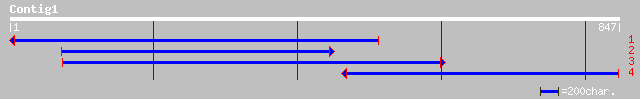
ref|NP_064043.1| orf112a [Beta vulgaris] gi|9049343|dbj|BAA99353... 100 5e-20
emb|CAA10612.1| ribosomal protein S14 [Pisum sativum] 67 3e-10
sp|P14875|RT14_OENBE MITOCHONDRIAL RIBOSOMAL PROTEIN S14 gi|1319... 67 3e-10
pir||R3OB4B ribosomal protein S14 - evening primrose mitochondrion 67 4e-10
sp|P49387|RT14_BRANA MITOCHONDRIAL RIBOSOMAL PROTEIN S14 gi|4869... 63 5e-09
>ref|NP_064043.1| orf112a [Beta vulgaris] gi|9049343|dbj|BAA99353.1| orf112a [Beta
vulgaris]
Length = 112
Score = 99.8 bits (247), Expect = 5e-20
Identities = 46/57 (80%), Positives = 51/57 (88%)
Frame = +1
Query: 169 PLPAGSGPKRPNGVIPYLGSEMLTGQESKWGTYLPPVSISCQVCSPDIDYVQGSTLG 339
P+PAGS ++ NG+IPYL SEMLTG+ESKWGT LPPVSISCQVC PDIDYVQGSTLG
Sbjct: 49 PIPAGSLTEKTNGLIPYLESEMLTGRESKWGTSLPPVSISCQVCFPDIDYVQGSTLG 105
>emb|CAA10612.1| ribosomal protein S14 [Pisum sativum]
Length = 100
Score = 67.0 bits (162), Expect = 3e-10
Identities = 32/33 (96%), Positives = 32/33 (96%)
Frame = +3
Query: 3 RPRSVYEFFRISRIVFRGLASRGSLMGIKKSSW 101
RPRSVYEFFRISRIVFRGLASRG LMGIKKSSW
Sbjct: 68 RPRSVYEFFRISRIVFRGLASRGPLMGIKKSSW 100
>sp|P14875|RT14_OENBE MITOCHONDRIAL RIBOSOMAL PROTEIN S14 gi|13194|emb|CAA34891.1|
ribosomal protein S14 (AA 1-99) [Oenothera berteriana]
Length = 99
Score = 67.0 bits (162), Expect = 3e-10
Identities = 32/33 (96%), Positives = 32/33 (96%)
Frame = +3
Query: 3 RPRSVYEFFRISRIVFRGLASRGSLMGIKKSSW 101
RPRSVYEFFRISRIVFRGLASRG LMGIKKSSW
Sbjct: 67 RPRSVYEFFRISRIVFRGLASRGPLMGIKKSSW 99
>pir||R3OB4B ribosomal protein S14 - evening primrose mitochondrion
Length = 99
Score = 66.6 bits (161), Expect = 4e-10
Identities = 32/33 (96%), Positives = 32/33 (96%)
Frame = +3
Query: 3 RPRSVYEFFRISRIVFRGLASRGSLMGIKKSSW 101
RPRSVYEFFRISRIVFRGLASRG LMGIKKSSW
Sbjct: 67 RPRSVYEFFRISRIVFRGLASRGLLMGIKKSSW 99
>sp|P49387|RT14_BRANA MITOCHONDRIAL RIBOSOMAL PROTEIN S14 gi|486984|pir||S36916 ribosomal
protein S14 - rape mitochondrion
gi|1524187|emb|CAA45191.1| ribosomal protein S14
[Brassica napus]
Length = 100
Score = 63.2 bits (152), Expect = 5e-09
Identities = 31/33 (93%), Positives = 31/33 (93%)
Frame = +3
Query: 3 RPRSVYEFFRISRIVFRGLASRGSLMGIKKSSW 101
RPRSV EFFRI RIVFRGLASRGSLMGIKKSSW
Sbjct: 68 RPRSVSEFFRIYRIVFRGLASRGSLMGIKKSSW 100
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 783,717,526
Number of Sequences: 1393205
Number of extensions: 18858813
Number of successful extensions: 48475
Number of sequences better than 10.0: 147
Number of HSP's better than 10.0 without gapping: 45407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48424
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 44316199683
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)