Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004925A_C01 KMC004925A_c01
(730 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
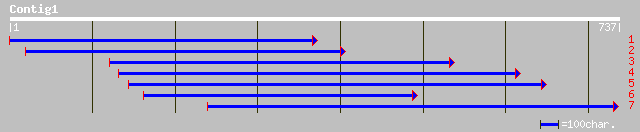
ref|NP_193847.2| putative protein; protein id: At4g21150.1, supp... 194 1e-48
dbj|BAB90282.1| P0470A12.7 [Oryza sativa (japonica cultivar-group)] 192 4e-48
pir||T04946 hypothetical protein F7J7.90 - Arabidopsis thaliana ... 141 1e-32
gb|AAH46052.1| Similar to ribophorin II [Danio rerio] 85 9e-16
gb|AAH46727.1| Similar to ribophorin II [Xenopus laevis] 84 3e-15
>ref|NP_193847.2| putative protein; protein id: At4g21150.1, supported by cDNA:
gi_16604453, supported by cDNA: gi_18958021 [Arabidopsis
thaliana] gi|16604454|gb|AAL24233.1| AT4g21150/F7J7_90
[Arabidopsis thaliana] gi|18958022|gb|AAL79584.1|
AT4g21150/F7J7_90 [Arabidopsis thaliana]
Length = 691
Score = 194 bits (492), Expect = 1e-48
Identities = 97/129 (75%), Positives = 108/129 (83%)
Frame = -2
Query: 729 PTPPVDPYSRYGPKAEINHIFRVPEKRPAQELSLAFLGLILLPFIGFLVGLLRLGVNLKN 550
P +PYSRYGPKAEI+HIFR+PEK PA++LSL FLG+I+LPFIGFL+GL RLGVN+K+
Sbjct: 563 PLQSTEPYSRYGPKAEISHIFRIPEKLPAKQLSLVFLGVIVLPFIGFLIGLTRLGVNIKS 622
Query: 549 FPSSTVPATFAILFHGGIAAVLLLYVLFWLKLDLFTTLKALGFLGAFLLFVGHRILSHLA 370
FPSST A A+LFH GI AVLLLYVLFWLKLDLFTTLKAL LG FLLFVGHR LS LA
Sbjct: 623 FPSSTGSAISALLFHCGIGAVLLLYVLFWLKLDLFTTLKALSLLGVFLLFVGHRTLSQLA 682
Query: 369 HTSAKLKSA 343
S KLKSA
Sbjct: 683 SASNKLKSA 691
>dbj|BAB90282.1| P0470A12.7 [Oryza sativa (japonica cultivar-group)]
Length = 689
Score = 192 bits (488), Expect = 4e-48
Identities = 95/132 (71%), Positives = 108/132 (80%), Gaps = 3/132 (2%)
Frame = -2
Query: 729 PTPP---VDPYSRYGPKAEINHIFRVPEKRPAQELSLAFLGLILLPFIGFLVGLLRLGVN 559
P PP VDP+S++GPK EI+HIFR PEKRP +ELS AF GL LLP +GFL+GL+RLGVN
Sbjct: 558 PKPPAQAVDPFSKFGPKKEISHIFRSPEKRPPKELSFAFTGLTLLPIVGFLIGLMRLGVN 617
Query: 558 LKNFPSSTVPATFAILFHGGIAAVLLLYVLFWLKLDLFTTLKALGFLGAFLLFVGHRILS 379
LKNFPS PA FA LFH GI AVLLLYVLFW+KLDLFTTLK L FLG FL+FVGHR LS
Sbjct: 618 LKNFPSLPAPAAFASLFHAGIGAVLLLYVLFWIKLDLFTTLKYLSFLGVFLVFVGHRALS 677
Query: 378 HLAHTSAKLKSA 343
+L+ TSAK K+A
Sbjct: 678 YLSSTSAKQKTA 689
>pir||T04946 hypothetical protein F7J7.90 - Arabidopsis thaliana
gi|2911072|emb|CAA17534.1| putative protein [Arabidopsis
thaliana] gi|7268912|emb|CAB79115.1| putative protein
[Arabidopsis thaliana]
Length = 609
Score = 141 bits (355), Expect = 1e-32
Identities = 68/98 (69%), Positives = 79/98 (80%)
Frame = -2
Query: 729 PTPPVDPYSRYGPKAEINHIFRVPEKRPAQELSLAFLGLILLPFIGFLVGLLRLGVNLKN 550
P +PYSRYGPKAEI+HIFR+PEK PA++LSL FLG+I+LPFIGFL+GL RLGVN+K+
Sbjct: 509 PLQSTEPYSRYGPKAEISHIFRIPEKLPAKQLSLVFLGVIVLPFIGFLIGLTRLGVNIKS 568
Query: 549 FPSSTVPATFAILFHGGIAAVLLLYVLFWLKLDLFTTL 436
FPSST A A+LFH GI AVLLLYVLFWLK L
Sbjct: 569 FPSSTGSAISALLFHCGIGAVLLLYVLFWLKASFHNRL 606
>gb|AAH46052.1| Similar to ribophorin II [Danio rerio]
Length = 630
Score = 85.1 bits (209), Expect = 9e-16
Identities = 47/110 (42%), Positives = 66/110 (59%)
Frame = -2
Query: 699 YGPKAEINHIFRVPEKRPAQELSLAFLGLILLPFIGFLVGLLRLGVNLKNFPSSTVPATF 520
Y PK EI H+FR PEK+P +S F L+L PF+ L+ +LG N+ NF S P+T
Sbjct: 518 YIPKPEIKHLFREPEKKPPTVVSNTFTALVLSPFLLLLILWFKLGANISNFTLS--PST- 574
Query: 519 AILFHGGIAAVLLLYVLFWLKLDLFTTLKALGFLGAFLLFVGHRILSHLA 370
ILFH G A +L L ++W L++F TLK L +G+ G+R+L+ A
Sbjct: 575 -ILFHIGHATMLGLMYVYWTHLNMFQTLKYLAIIGSLTFLAGNRMLAQKA 623
>gb|AAH46727.1| Similar to ribophorin II [Xenopus laevis]
Length = 631
Score = 83.6 bits (205), Expect = 3e-15
Identities = 45/117 (38%), Positives = 68/117 (57%)
Frame = -2
Query: 720 PVDPYSRYGPKAEINHIFRVPEKRPAQELSLAFLGLILLPFIGFLVGLLRLGVNLKNFPS 541
P+ + + PK +I H+FR PEKRP +S F LIL P + + +++G N+ N
Sbjct: 513 PIQSKNLFIPKPDIQHLFREPEKRPPTVVSNTFTALILAPLLLLFILWIKIGANVSNISF 572
Query: 540 STVPATFAILFHGGIAAVLLLYVLFWLKLDLFTTLKALGFLGAFLLFVGHRILSHLA 370
S P+T I+FH G A +L L ++W +L++F TLK L LG G+R+L+H A
Sbjct: 573 S--PST--IIFHLGHATMLGLMYVYWTQLNMFQTLKYLALLGTITFLAGNRMLAHKA 625
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 603,691,348
Number of Sequences: 1393205
Number of extensions: 13256800
Number of successful extensions: 32219
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 31281
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32191
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34343566934
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)