Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004920A_C01 KMC004920A_c01
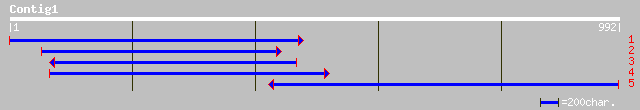
(990 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM13186.1| putative receptor-like protein kinase [Arabidopsi... 434 e-120
ref|NP_180000.2| leucine-rich repeat transmembrane protein kinas... 434 e-120
pir||B84634 probable receptor-like protein kinase [imported] - A... 434 e-120
gb|AAF73754.1|AF149037_1 receptor-like protein kinase [Prunus du... 395 e-109
ref|NP_200623.1| leucine-rich repeat transmembrane protein kinas... 241 1e-62
>gb|AAM13186.1| putative receptor-like protein kinase [Arabidopsis thaliana]
Length = 853
Score = 434 bits (1115), Expect = e-120
Identities = 206/258 (79%), Positives = 230/258 (88%)
Frame = -1
Query: 990 FLGRIKHPNLVLLTGYCLAGDQRIAIYDYMENGNLQNLLYDLPLGVLHSTDDWSTDTWEE 811
FLGRIKHPNLV LTGYC+AGDQRIAIY+YMENGNLQNLL+DLP GV +TDDW+TDTWEE
Sbjct: 593 FLGRIKHPNLVPLTGYCIAGDQRIAIYEYMENGNLQNLLHDLPFGV-QTTDDWTTDTWEE 651
Query: 810 PDNNGIQNAGSEGLLTTWSFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 631
+NG QN G+EG + TW FRHKIALGTARALAFLHHGCSPPIIHR VKASSVYLD + E
Sbjct: 652 ETDNGTQNIGTEGPVATWRFRHKIALGTARALAFLHHGCSPPIIHRDVKASSVYLDQNWE 711
Query: 630 PRLSDFGLAKIFGSGLDEEIARGSPGYDPPEFTQPDFDTPTTKSDVYCFGVVLFELLTGK 451
PRLSDFGLAK+FG+GLD+EI GSPGY PPEF QP+ + PT KSDVYCFGVVLFEL+TGK
Sbjct: 712 PRLSDFGLAKVFGNGLDDEIIHGSPGYLPPEFLQPEHELPTPKSDVYCFGVVLFELMTGK 771
Query: 450 KPVEDDYHDDKEETLVSWVRGLVRKNQTSRAIDPKIRDTGPDEQMEEALKIGYLCTADLP 271
KP+EDDY D+K+ LVSWVR LVRKNQ S+AIDPKI++TG +EQMEEALKIGYLCTADLP
Sbjct: 772 KPIEDDYLDEKDTNLVSWVRSLVRKNQASKAIDPKIQETGSEEQMEEALKIGYLCTADLP 831
Query: 270 FKRPTMQQIVGLLKDIEP 217
KRP+MQQ+VGLLKDIEP
Sbjct: 832 SKRPSMQQVVGLLKDIEP 849
>ref|NP_180000.2| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At2g24230.1, supported by cDNA: gi_20260575
[Arabidopsis thaliana]
Length = 853
Score = 434 bits (1115), Expect = e-120
Identities = 206/258 (79%), Positives = 230/258 (88%)
Frame = -1
Query: 990 FLGRIKHPNLVLLTGYCLAGDQRIAIYDYMENGNLQNLLYDLPLGVLHSTDDWSTDTWEE 811
FLGRIKHPNLV LTGYC+AGDQRIAIY+YMENGNLQNLL+DLP GV +TDDW+TDTWEE
Sbjct: 593 FLGRIKHPNLVPLTGYCIAGDQRIAIYEYMENGNLQNLLHDLPFGV-QTTDDWTTDTWEE 651
Query: 810 PDNNGIQNAGSEGLLTTWSFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 631
+NG QN G+EG + TW FRHKIALGTARALAFLHHGCSPPIIHR VKASSVYLD + E
Sbjct: 652 ETDNGTQNIGTEGPVATWRFRHKIALGTARALAFLHHGCSPPIIHRDVKASSVYLDQNWE 711
Query: 630 PRLSDFGLAKIFGSGLDEEIARGSPGYDPPEFTQPDFDTPTTKSDVYCFGVVLFELLTGK 451
PRLSDFGLAK+FG+GLD+EI GSPGY PPEF QP+ + PT KSDVYCFGVVLFEL+TGK
Sbjct: 712 PRLSDFGLAKVFGNGLDDEIIHGSPGYLPPEFLQPEHELPTPKSDVYCFGVVLFELMTGK 771
Query: 450 KPVEDDYHDDKEETLVSWVRGLVRKNQTSRAIDPKIRDTGPDEQMEEALKIGYLCTADLP 271
KP+EDDY D+K+ LVSWVR LVRKNQ S+AIDPKI++TG +EQMEEALKIGYLCTADLP
Sbjct: 772 KPIEDDYLDEKDTNLVSWVRSLVRKNQASKAIDPKIQETGSEEQMEEALKIGYLCTADLP 831
Query: 270 FKRPTMQQIVGLLKDIEP 217
KRP+MQQ+VGLLKDIEP
Sbjct: 832 SKRPSMQQVVGLLKDIEP 849
>pir||B84634 probable receptor-like protein kinase [imported] - Arabidopsis
thaliana gi|4115383|gb|AAD03384.1| putative receptor-like
protein kinase [Arabidopsis thaliana]
Length = 809
Score = 434 bits (1115), Expect = e-120
Identities = 206/258 (79%), Positives = 230/258 (88%)
Frame = -1
Query: 990 FLGRIKHPNLVLLTGYCLAGDQRIAIYDYMENGNLQNLLYDLPLGVLHSTDDWSTDTWEE 811
FLGRIKHPNLV LTGYC+AGDQRIAIY+YMENGNLQNLL+DLP GV +TDDW+TDTWEE
Sbjct: 549 FLGRIKHPNLVPLTGYCIAGDQRIAIYEYMENGNLQNLLHDLPFGV-QTTDDWTTDTWEE 607
Query: 810 PDNNGIQNAGSEGLLTTWSFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 631
+NG QN G+EG + TW FRHKIALGTARALAFLHHGCSPPIIHR VKASSVYLD + E
Sbjct: 608 ETDNGTQNIGTEGPVATWRFRHKIALGTARALAFLHHGCSPPIIHRDVKASSVYLDQNWE 667
Query: 630 PRLSDFGLAKIFGSGLDEEIARGSPGYDPPEFTQPDFDTPTTKSDVYCFGVVLFELLTGK 451
PRLSDFGLAK+FG+GLD+EI GSPGY PPEF QP+ + PT KSDVYCFGVVLFEL+TGK
Sbjct: 668 PRLSDFGLAKVFGNGLDDEIIHGSPGYLPPEFLQPEHELPTPKSDVYCFGVVLFELMTGK 727
Query: 450 KPVEDDYHDDKEETLVSWVRGLVRKNQTSRAIDPKIRDTGPDEQMEEALKIGYLCTADLP 271
KP+EDDY D+K+ LVSWVR LVRKNQ S+AIDPKI++TG +EQMEEALKIGYLCTADLP
Sbjct: 728 KPIEDDYLDEKDTNLVSWVRSLVRKNQASKAIDPKIQETGSEEQMEEALKIGYLCTADLP 787
Query: 270 FKRPTMQQIVGLLKDIEP 217
KRP+MQQ+VGLLKDIEP
Sbjct: 788 SKRPSMQQVVGLLKDIEP 805
>gb|AAF73754.1|AF149037_1 receptor-like protein kinase [Prunus dulcis]
Length = 221
Score = 395 bits (1016), Expect = e-109
Identities = 188/222 (84%), Positives = 207/222 (92%)
Frame = -1
Query: 885 QNLLYDLPLGVLHSTDDWSTDTWEEPDNNGIQNAGSEGLLTTWSFRHKIALGTARALAFL 706
QNLLYDLPLGV +T+DWSTDTWEE DNNGIQN GSEGLLTTW FRHKIALGTARALAFL
Sbjct: 1 QNLLYDLPLGV-QTTEDWSTDTWEEDDNNGIQNVGSEGLLTTWRFRHKIALGTARALAFL 59
Query: 705 HHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIARGSPGYDPPEFTQP 526
HHGCSPPIIHR VKASSVYLDY+LEPRLSDFGLAKIFG+GLDEEI+RGSPGY PPEF+QP
Sbjct: 60 HHGCSPPIIHRDVKASSVYLDYNLEPRLSDFGLAKIFGNGLDEEISRGSPGYLPPEFSQP 119
Query: 525 DFDTPTTKSDVYCFGVVLFELLTGKKPVEDDYHDDKEETLVSWVRGLVRKNQTSRAIDPK 346
++DTPT KSDVYCFGVVLFEL+TGKKP+ DDY ++K+ TLVSWVRGLV+KN+ + AIDPK
Sbjct: 120 EYDTPTPKSDVYCFGVVLFELITGKKPIGDDYPEEKDATLVSWVRGLVKKNRGASAIDPK 179
Query: 345 IRDTGPDEQMEEALKIGYLCTADLPFKRPTMQQIVGLLKDIE 220
IRDTGPD+QMEEALKIGYLCTADLP KRP+M QIVGLLKD+E
Sbjct: 180 IRDTGPDDQMEEALKIGYLCTADLPLKRPSMHQIVGLLKDME 221
>ref|NP_200623.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At5g58150.1, supported by cDNA: gi_20268687
[Arabidopsis thaliana] gi|8777316|dbj|BAA96906.1|
receptor-like protein kinase [Arabidopsis thaliana]
gi|20268688|gb|AAM14048.1| putative receptor protein
kinase [Arabidopsis thaliana] gi|21689849|gb|AAM67568.1|
putative receptor protein kinase [Arabidopsis thaliana]
Length = 785
Score = 241 bits (615), Expect = 1e-62
Identities = 136/259 (52%), Positives = 169/259 (64%), Gaps = 2/259 (0%)
Frame = -1
Query: 987 LGRIKHPNLVLLTGYCLAGDQRIAIYDYMENGNLQNLLYDLPLGVLHSTDDWSTDTWEEP 808
L RI HPNL L GYC+A +QRIAIY+ ++ NLQ+LL+
Sbjct: 569 LARINHPNLFPLCGYCIATEQRIAIYEDLDMVNLQSLLH--------------------- 607
Query: 807 DNNGIQNAGSEGLLTTWSFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEP 628
NNG +A W RHKIALGTARALAFLHHGC PP++H VKA+++ LD EP
Sbjct: 608 -NNGDDSA-------PWRLRHKIALGTARALAFLHHGCIPPMVHGEVKAATILLDSSQEP 659
Query: 627 RLSDFGLAKIFGSGLDEEI--ARGSPGYDPPEFTQPDFDTPTTKSDVYCFGVVLFELLTG 454
RL+DFGL K+ LDE+ + GY PPE Q +PT +SDVY FGVVL EL++G
Sbjct: 660 RLADFGLVKL----LDEQFPGSESLDGYTPPE--QERNASPTLESDVYSFGVVLLELVSG 713
Query: 453 KKPVEDDYHDDKEETLVSWVRGLVRKNQTSRAIDPKIRDTGPDEQMEEALKIGYLCTADL 274
KKP D LV+WVRGLVR+ Q RAIDP +++T P++++ EA+KIGYLCTADL
Sbjct: 714 KKPEGD---------LVNWVRGLVRQGQGLRAIDPTMQETVPEDEIAEAVKIGYLCTADL 764
Query: 273 PFKRPTMQQIVGLLKDIEP 217
P+KRPTMQQ+VGLLKDI P
Sbjct: 765 PWKRPTMQQVVGLLKDISP 783
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 883,487,367
Number of Sequences: 1393205
Number of extensions: 19993893
Number of successful extensions: 69296
Number of sequences better than 10.0: 7514
Number of HSP's better than 10.0 without gapping: 56359
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 62385
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 56566024535
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)