Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004894A_C01 KMC004894A_c01
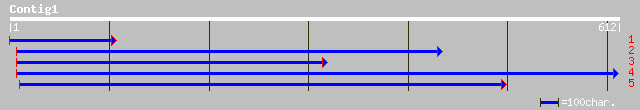
(611 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_192248.1| leucine-rich repeat transmembrane protein kinas... 58 1e-07
ref|NP_565489.1| putative LRR receptor protein kinase; protein i... 56 4e-07
pir||B84594 probable LRR receptor protein kinase [imported] - Ar... 56 4e-07
ref|NP_178019.1| leucine-rich repeat transmembrane protein kinas... 49 7e-05
ref|NP_193944.1| serine/threonine protein kinase like protein; p... 47 2e-04
>ref|NP_192248.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At4g03390.1 [Arabidopsis thaliana]
gi|25407189|pir||A85043 probable LRR receptor-like
protein kinase [imported] - Arabidopsis thaliana
gi|4262167|gb|AAD14467.1| putative LRR receptor-linked
protein kinase [Arabidopsis thaliana]
gi|7270209|emb|CAB77824.1| putative LRR receptor-like
protein kinase [Arabidopsis thaliana]
Length = 754
Score = 57.8 bits (138), Expect = 1e-07
Identities = 29/35 (82%), Positives = 31/35 (87%)
Frame = -3
Query: 609 STFADIISRCVQSEPEFRPAMSEVVLYLLNMIRKE 505
S FADIISRCVQSEPEFRP MSEVV LL+MIR+E
Sbjct: 710 SHFADIISRCVQSEPEFRPLMSEVVQDLLDMIRRE 744
>ref|NP_565489.1| putative LRR receptor protein kinase; protein id: At2g20850.1,
supported by cDNA: gi_15810274 [Arabidopsis thaliana]
gi|15810275|gb|AAL07025.1| putative LRR receptor protein
kinase [Arabidopsis thaliana] gi|20197699|gb|AAD20910.3|
putative LRR receptor protein kinase [Arabidopsis
thaliana] gi|25054921|gb|AAN71938.1| putative LRR
receptor protein kinase [Arabidopsis thaliana]
Length = 772
Score = 55.8 bits (133), Expect = 4e-07
Identities = 26/40 (65%), Positives = 34/40 (85%)
Frame = -3
Query: 609 STFADIISRCVQSEPEFRPAMSEVVLYLLNMIRKESQQNE 490
S FAD+ISRCVQSEPE+RP MSEVV L +MI++E ++N+
Sbjct: 722 SHFADVISRCVQSEPEYRPLMSEVVQDLSDMIQREHRRND 761
>pir||B84594 probable LRR receptor protein kinase [imported] - Arabidopsis
thaliana
Length = 767
Score = 55.8 bits (133), Expect = 4e-07
Identities = 26/40 (65%), Positives = 34/40 (85%)
Frame = -3
Query: 609 STFADIISRCVQSEPEFRPAMSEVVLYLLNMIRKESQQNE 490
S FAD+ISRCVQSEPE+RP MSEVV L +MI++E ++N+
Sbjct: 717 SHFADVISRCVQSEPEYRPLMSEVVQDLSDMIQREHRRND 756
>ref|NP_178019.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At1g78980.1 [Arabidopsis thaliana]
Length = 693
Score = 48.5 bits (114), Expect = 7e-05
Identities = 23/38 (60%), Positives = 29/38 (75%)
Frame = -3
Query: 609 STFADIISRCVQSEPEFRPAMSEVVLYLLNMIRKESQQ 496
S FADII+ CVQ EPEFRP MSEVV L+ M+++ S +
Sbjct: 639 SRFADIIALCVQVEPEFRPPMSEVVEALVRMVQRSSMK 676
>ref|NP_193944.1| serine/threonine protein kinase like protein; protein id:
At4g22130.1 [Arabidopsis thaliana]
gi|11259536|pir||T49120 serine/threonine protein kinase
like protein - Arabidopsis thaliana
gi|2961358|emb|CAA18116.1| serine/threonine protein
kinase like protein [Arabidopsis thaliana]
gi|7269058|emb|CAB79168.1| serine/threonine protein
kinase like protein [Arabidopsis thaliana]
Length = 338
Score = 47.4 bits (111), Expect = 2e-04
Identities = 21/36 (58%), Positives = 28/36 (77%)
Frame = -3
Query: 609 STFADIISRCVQSEPEFRPAMSEVVLYLLNMIRKES 502
S FADII+ C+Q EPEFRP MSEVV L+ ++++ S
Sbjct: 277 SRFADIIALCIQPEPEFRPPMSEVVQQLVRLVQRAS 312
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,216,973
Number of Sequences: 1393205
Number of extensions: 9684988
Number of successful extensions: 20764
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 20322
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20763
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)