Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
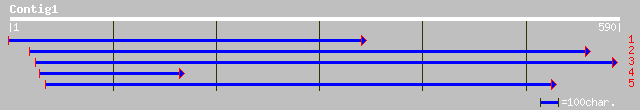
Query= KMC004877A_C01 KMC004877A_c01
(583 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_608341.1| CG14223-PA [Drosophila melanogaster] gi|7293625... 37 0.23
ref|NP_296373.1| F-box only protein 21 isoform 1; F-box protein ... 33 2.0
ref|NP_055817.1| F-box only protein 21 isoform 2; F-box protein ... 33 2.0
dbj|BAC36377.1| unnamed protein product [Mus musculus] 33 2.6
ref|NP_663539.1| similar to F-box only protein 21, isoform 1; F-... 33 2.6
>ref|NP_608341.1| CG14223-PA [Drosophila melanogaster] gi|7293625|gb|AAF48997.1|
CG14223-PA [Drosophila melanogaster]
Length = 581
Score = 36.6 bits (83), Expect = 0.23
Identities = 24/70 (34%), Positives = 33/70 (46%), Gaps = 3/70 (4%)
Frame = +1
Query: 328 SNKNRPQKIQHHHLQQAYHVCWN-WHTHG*ASHCLWLQLTCTQSHL--HVLNLPLSSDNP 498
S++ + Q+ QHHH QQ +H N H H +H T S L + N S NP
Sbjct: 203 SHQQQQQQHQHHHQQQQHHNHHNHHHQHQQTNHAQAQSQTHNLSQLPQNYRNFSHPSTNP 262
Query: 499 LKHKNPYQSP 528
++HK P P
Sbjct: 263 VRHKLPKSGP 272
>ref|NP_296373.1| F-box only protein 21 isoform 1; F-box protein 21 [Homo sapiens]
Length = 628
Score = 33.5 bits (75), Expect = 2.0
Identities = 19/56 (33%), Positives = 29/56 (50%), Gaps = 3/56 (5%)
Frame = +3
Query: 309 SAEDVSLEQEPTPENTAPPSSTGISCMLELAYPWLSFTL---SLAAIDLHSIASTC 467
+A D ++E P A P G+SC++ L L + L SL A D+ ++STC
Sbjct: 4 AAVDSAMEVVPALAEEAAPEVAGLSCLVNLPGEVLEYILCCGSLTAADIGRVSSTC 59
>ref|NP_055817.1| F-box only protein 21 isoform 2; F-box protein 21 [Homo sapiens]
gi|4240239|dbj|BAA74898.1| KIAA0875 protein [Homo
sapiens] gi|6164745|gb|AAF04522.1|AF174601_1 F-box
protein Fbx21 [Homo sapiens]
Length = 621
Score = 33.5 bits (75), Expect = 2.0
Identities = 19/56 (33%), Positives = 29/56 (50%), Gaps = 3/56 (5%)
Frame = +3
Query: 309 SAEDVSLEQEPTPENTAPPSSTGISCMLELAYPWLSFTL---SLAAIDLHSIASTC 467
+A D ++E P A P G+SC++ L L + L SL A D+ ++STC
Sbjct: 4 AAVDSAMEVVPALAEEAAPEVAGLSCLVNLPGEVLEYILCCGSLTAADIGRVSSTC 59
>dbj|BAC36377.1| unnamed protein product [Mus musculus]
Length = 242
Score = 33.1 bits (74), Expect = 2.6
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Frame = +3
Query: 312 AEDVSLEQEPTPENTAPPSSTGISCMLELAYPWLSFTL---SLAAIDLHSIASTC 467
A D ++E P A +TG SC+++L L + L SL A+D+ ++STC
Sbjct: 5 AGDSAMEVVPALAEEAAAEATGPSCLVQLPGEVLEYILCSGSLTALDIGRVSSTC 59
>ref|NP_663539.1| similar to F-box only protein 21, isoform 1; F-box protein 21 [Mus
musculus] gi|18256900|gb|AAH21871.1| Similar to F-box
only protein 21 [Mus musculus]
gi|26340462|dbj|BAC33894.1| unnamed protein product [Mus
musculus]
Length = 627
Score = 33.1 bits (74), Expect = 2.6
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Frame = +3
Query: 312 AEDVSLEQEPTPENTAPPSSTGISCMLELAYPWLSFTL---SLAAIDLHSIASTC 467
A D ++E P A +TG SC+++L L + L SL A+D+ ++STC
Sbjct: 5 AGDSAMEVVPALAEEAAAEATGPSCLVQLPGEVLEYILCSGSLTALDIGRVSSTC 59
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 474,979,346
Number of Sequences: 1393205
Number of extensions: 10253886
Number of successful extensions: 26287
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 25266
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26249
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)