Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004862A_C01 KMC004862A_c01
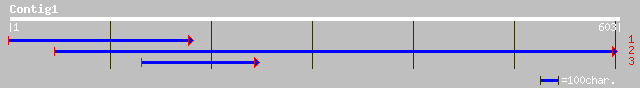
(603 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_180055.1| putative Rieske iron-sulfur protein; protein id... 35 0.57
pir||T06499 Rieske [2Fe-2S] iron-sulfur protein tic55 - garden p... 35 0.57
ref|NP_650849.1| CG4462-PA [Drosophila melanogaster] gi|7300568|... 33 2.8
gb|AAM83014.1| polymerase [Hepatitis B virus] 32 4.8
pir||S63539 GABA/beta-alanine transporter - marbled electric ray... 32 4.8
>ref|NP_180055.1| putative Rieske iron-sulfur protein; protein id: At2g24820.1,
supported by cDNA: 29774. [Arabidopsis thaliana]
gi|25412251|pir||H84640 probable Rieske iron-sulfur
protein [imported] - Arabidopsis thaliana
gi|4559369|gb|AAD23030.1| putative Rieske iron-sulfur
protein [Arabidopsis thaliana]
Length = 539
Score = 35.4 bits (80), Expect = 0.57
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = -3
Query: 301 YEWTEKWYHLYLT*LGNNDAALGVSGY 221
Y+WTE+WY LYLT DA LG++ Y
Sbjct: 82 YDWTEEWYPLYLTKNVPEDAPLGLTVY 108
>pir||T06499 Rieske [2Fe-2S] iron-sulfur protein tic55 - garden pea
gi|2764524|emb|CAA04157.1| Rieske iron-sulfur protein
Tic55 [Pisum sativum]
Length = 553
Score = 35.4 bits (80), Expect = 0.57
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = -3
Query: 301 YEWTEKWYHLYLT*LGNNDAALGVSGY 221
Y+WTE+WY LYLT +DA LG+ Y
Sbjct: 97 YDWTEEWYPLYLTKNVPHDAPLGLKVY 123
>ref|NP_650849.1| CG4462-PA [Drosophila melanogaster] gi|7300568|gb|AAF55720.1|
CG4462-PA [Drosophila melanogaster]
Length = 573
Score = 33.1 bits (74), Expect = 2.8
Identities = 33/121 (27%), Positives = 51/121 (41%), Gaps = 13/121 (10%)
Frame = -3
Query: 373 SSCSSFSAVPEH----FQFNLRTN*KIYYEWTEKWYHLYLT*LG---NNDAALGV---SG 224
S C F+ V QFNL I+ WT+ W HL+ +G LG+ S
Sbjct: 121 SECEQFNYVSSFDSLIMQFNLVCLRDIFVAWTQYW-HLFGVLVGGVMGTKMMLGISPRST 179
Query: 223 YRVFFFARILCGVSSVVNRN-KVKALASCYGVFVCLLLIIS--VCFPDLNHGSTKLMKLM 53
Y V A+ILCGV + R+ + C C ++ + F D+ G ++ ++
Sbjct: 180 YCVGAVAQILCGVVTGYARDFSLHCAFRCLSAVCCAIMFTAGQAIFADITAGMHRIGAII 239
Query: 52 L 50
L
Sbjct: 240 L 240
>gb|AAM83014.1| polymerase [Hepatitis B virus]
Length = 226
Score = 32.3 bits (72), Expect = 4.8
Identities = 17/54 (31%), Positives = 28/54 (51%)
Frame = +3
Query: 426 NMRVSCTKQQYQLGICIAIRESSTSTLGKEIVDLSQKKKTDTRVSMVKFVIGGL 587
N+R+S T + G+ + + +T + +VD SQ + TRVS KF + L
Sbjct: 4 NIRISRTPARVTGGVFLVDKNPHNTTESRLVVDFSQFSRGSTRVSWPKFAVPNL 57
>pir||S63539 GABA/beta-alanine transporter - marbled electric ray
gi|1171630|emb|CAA60635.1| GABA/beta-alanine transporter
[Torpedo marmorata]
Length = 622
Score = 32.3 bits (72), Expect = 4.8
Identities = 14/27 (51%), Positives = 19/27 (69%)
Frame = +1
Query: 184 TPHIRFWQKRILGTHLHRGLHHCSRVR 264
+P I FW++R+LG L RG+ H RVR
Sbjct: 189 SPVIEFWERRVLG--LSRGIEHIGRVR 213
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,678,358
Number of Sequences: 1393205
Number of extensions: 11129197
Number of successful extensions: 26262
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 25604
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26254
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)