Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004857A_C01 KMC004857A_c01
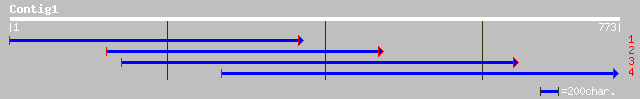
(772 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_176191.1| chromaffin granule ATPase II homolog, putative;... 219 3e-56
sp|Q9XIE6|ALA3_ARATH Potential phospholipid-transporting ATPase ... 60 3e-08
emb|CAC83304.1| putative ATPase [Pinus pinaster] 47 3e-04
ref|NP_596486.1| putative calcium-transporting atpase [Schizosac... 42 0.010
ref|NP_009376.1| P-type ATPase, potential aminophospholipid tran... 42 0.013
>ref|NP_176191.1| chromaffin granule ATPase II homolog, putative; protein id:
At1g59820.1, supported by cDNA: gi_20147218 [Arabidopsis
thaliana] gi|20147219|gb|AAM10325.1| At1g59820/F23H11_14
[Arabidopsis thaliana]
Length = 1213
Score = 219 bits (559), Expect = 3e-56
Identities = 103/132 (78%), Positives = 119/132 (90%), Gaps = 1/132 (0%)
Frame = -2
Query: 768 ENIYFVIYVLMSTLYFYLTLLLVPVSALFCDFVYQGVQRWFFPYDYQIIQEIHRHELDNT 589
EN+YFVIYVLMST YFY TLLLVP+ +L DF++QGV+RWFFPYDYQI+QEIHRHE D +
Sbjct: 1079 ENVYFVIYVLMSTFYFYFTLLLVPIVSLLGDFIFQGVERWFFPYDYQIVQEIHRHESDAS 1138
Query: 588 GRAQL-LESQLTPAEARSYAISQLPREISKHTGFAFDSPGYESFFAAQLGVYAPPKAWDV 412
QL +E++LTP EARSYAISQLPRE+SKHTGFAFDSPGYESFFA+QLG+YAP KAWDV
Sbjct: 1139 KADQLEVENELTPQEARSYAISQLPRELSKHTGFAFDSPGYESFFASQLGIYAPQKAWDV 1198
Query: 411 ARRASMKSRPKI 376
ARRASM+SRPK+
Sbjct: 1199 ARRASMRSRPKV 1210
>sp|Q9XIE6|ALA3_ARATH Potential phospholipid-transporting ATPase 3 (Aminophospholipid
flippase 3) gi|25404236|pir||C96622 probable ATPase
F23H11.14 [imported] - Arabidopsis thaliana
gi|5080816|gb|AAD39325.1|AC007258_14 Putative ATPase
[Arabidopsis thaliana]
Length = 1123
Score = 60.1 bits (144), Expect = 3e-08
Identities = 26/36 (72%), Positives = 31/36 (85%)
Frame = -2
Query: 768 ENIYFVIYVLMSTLYFYLTLLLVPVSALFCDFVYQG 661
EN+YFVIYVLMST YFY TLLLVP+ +L DF++QG
Sbjct: 1079 ENVYFVIYVLMSTFYFYFTLLLVPIVSLLGDFIFQG 1114
>emb|CAC83304.1| putative ATPase [Pinus pinaster]
Length = 133
Score = 47.0 bits (110), Expect = 3e-04
Identities = 20/37 (54%), Positives = 31/37 (83%)
Frame = -2
Query: 771 QENIYFVIYVLMSTLYFYLTLLLVPVSALFCDFVYQG 661
Q ++Y+VI VL+ST YF+L+++LVP+ AL DF++QG
Sbjct: 97 QASVYYVICVLVSTWYFWLSIILVPLVALLGDFLFQG 133
>ref|NP_596486.1| putative calcium-transporting atpase [Schizosaccharomyces pombe]
gi|7492445|pir||T40737 probable calcium-transporting
atpase - fission yeast (Schizosaccharomyces pombe)
gi|3850108|emb|CAA21897.1| putative calcium-transporting
atpase [Schizosaccharomyces pombe]
Length = 1258
Score = 42.0 bits (97), Expect = 0.010
Identities = 19/54 (35%), Positives = 33/54 (60%)
Frame = -2
Query: 759 YFVIYVLMSTLYFYLTLLLVPVSALFCDFVYQGVQRWFFPYDYQIIQEIHRHEL 598
Y +I L L F+ +LL++P AL DFV++ R ++P +Y +QEI ++ +
Sbjct: 1147 YGIIPHLYGNLKFWASLLVLPTIALMRDFVWKYSSRMYYPEEYHYVQEIQKYNV 1200
>ref|NP_009376.1| P-type ATPase, potential aminophospholipid translocase; Drs2p
[Saccharomyces cerevisiae] gi|728905|sp|P39524|ATC3_YEAST
Potential phospholipid-transporting ATPase 1
gi|1078206|pir||S51995 probable ATPase (EC 3.6.1.-) DRS2
- yeast (Saccharomyces cerevisiae)
gi|171114|gb|AAA16891.1| ATPase gi|595560|gb|AAC05006.1|
Drs2p: Membrane spanning Ca-ATPase(P-type), member of the
cation transport(E1-E2) ATPase [Saccharomyces cerevisiae]
Length = 1355
Score = 41.6 bits (96), Expect = 0.013
Identities = 23/80 (28%), Positives = 45/80 (55%)
Frame = -2
Query: 723 FYLTLLLVPVSALFCDFVYQGVQRWFFPYDYQIIQEIHRHELDNTGRAQLLESQLTPAEA 544
F+LTL+++P+ AL DF+++ +R + P Y +IQE+ ++ + ++ R + + Q +
Sbjct: 1205 FWLTLIVLPIFALVRDFLWKYYKRMYEPETYHVIQEMQKYNISDS-RPHVQQFQNAIRKV 1263
Query: 543 RSYAISQLPREISKHTGFAF 484
R + + K GFAF
Sbjct: 1264 RQV------QRMKKQRGFAF 1277
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 665,386,890
Number of Sequences: 1393205
Number of extensions: 14714645
Number of successful extensions: 38801
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 37263
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38764
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37815044670
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)