Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004834A_C01 KMC004834A_c01
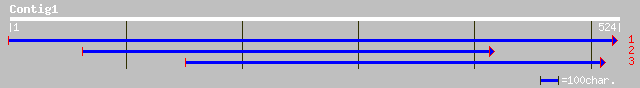
(524 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_013903.1| Hypothetical ORF; Ymr178wp [Saccharomyces cerev... 32 4.5
ref|NP_242281.1| BH1415~unknown conserved protein in others [Bac... 32 5.9
>ref|NP_013903.1| Hypothetical ORF; Ymr178wp [Saccharomyces cerevisiae]
gi|2497175|sp|Q03219|YM44_YEAST HYPOTHETICAL 31.1 KD
PROTEIN IN SIP18-SPT21 INTERGENIC REGION
gi|1078525|pir||S55125 hypothetical protein YMR178w -
yeast (Saccharomyces cerevisiae)
gi|854448|emb|CAA89911.1| unknown [Saccharomyces
cerevisiae]
Length = 274
Score = 32.0 bits (71), Expect = 4.5
Identities = 13/34 (38%), Positives = 21/34 (61%)
Frame = +1
Query: 241 FLSEHVTSAHISKQEKTIQTPNGKIREEVKVAEY 342
F+ H+T + ISK+ K IQ + K+ E +K+ Y
Sbjct: 190 FVRTHLTESQISKELKLIQDESTKVSEAIKIGSY 223
>ref|NP_242281.1| BH1415~unknown conserved protein in others [Bacillus halodurans]
gi|25489849|pir||G83826 hypothetical protein BH1415
[imported] - Bacillus halodurans (strain C-125)
gi|10174032|dbj|BAB05134.1| BH1415~unknown conserved
protein in others [Bacillus halodurans]
Length = 923
Score = 31.6 bits (70), Expect = 5.9
Identities = 18/68 (26%), Positives = 28/68 (40%)
Frame = +1
Query: 160 HFLILNRIQESHIYFNNPLDQPTIYM*FLSEHVTSAHISKQEKTIQTPNGKIREEVKVAE 339
H + I E + DQ + F+ EHV S + +K NG RE+++ A
Sbjct: 671 HTELQQAIHEQEMKLVTEADQIIVCSQFMKEHVQSLFVPNPDKVAVIANGVAREQIEAAR 730
Query: 340 YLTANTLN 363
T + N
Sbjct: 731 LQTISPEN 738
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 432,760,927
Number of Sequences: 1393205
Number of extensions: 9158174
Number of successful extensions: 21485
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 20854
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21474
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17019769648
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)