Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004832A_C01 KMC004832A_c01
(561 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
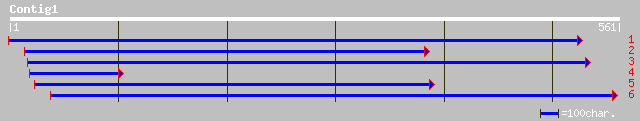
ref|NP_199845.1| DNA repair protein-like; protein id: At5g50340.... 169 3e-41
gb|ZP_00053991.1| hypothetical protein [Magnetospirillum magneto... 90 2e-17
ref|NP_710128.1| probable ATP-dependent protease [Shigella flexn... 90 2e-17
ref|NP_757320.1| DNA repair protein radA [Escherichia coli CFT07... 90 2e-17
ref|NP_418806.1| probable ATP-dependent protease [Escherichia co... 90 2e-17
>ref|NP_199845.1| DNA repair protein-like; protein id: At5g50340.1 [Arabidopsis
thaliana] gi|9758916|dbj|BAB09453.1| DNA repair
protein-like [Arabidopsis thaliana]
Length = 484
Score = 169 bits (427), Expect = 3e-41
Identities = 79/122 (64%), Positives = 105/122 (85%)
Frame = -2
Query: 560 MIISVLIKQAGLRLQEHAVFLNVVSGLTLAETAGDLAVAAAICSSCLELPIPNDIAFIGE 381
MII+VL+KQAGLR+QE+ +FLNV +G+ L+ETAGDLA+AAAICSS LE PIP+ +AFIGE
Sbjct: 361 MIIAVLMKQAGLRIQENGIFLNVANGMALSETAGDLAIAAAICSSFLEFPIPHGVAFIGE 420
Query: 380 IGLGGELRMVPRMEKRMYTVAKLGYRMCIVPKAAEKASGAEGLENMKVVGCRNLKDVINT 201
IGLGGE+R VPRMEKR+ TVAKLG+ C+VPK+ E++ A L+ ++++GC+NLK++IN
Sbjct: 421 IGLGGEVRTVPRMEKRVSTVAKLGFNKCVVPKSVEESLKALSLKEIEIIGCKNLKELINA 480
Query: 200 VF 195
VF
Sbjct: 481 VF 482
>gb|ZP_00053991.1| hypothetical protein [Magnetospirillum magnetotacticum]
Length = 381
Score = 90.1 bits (222), Expect = 2e-17
Identities = 50/121 (41%), Positives = 75/121 (61%), Gaps = 3/121 (2%)
Frame = -2
Query: 560 MIISVLIKQAGLRLQEHAVFLNVVSGLTLAETAGDLAVAAAICSSCLELPIPNDIAFIGE 381
M+++VL + GL L + V+LNV GL +AE A DLAVAAA+ SS ++P+P D+ GE
Sbjct: 255 MVLAVLDARCGLALSGNDVYLNVAGGLRIAEPAADLAVAAALVSSASDVPVPADMVVFGE 314
Query: 380 IGLGGELRMVPRMEKRMYTVAKLGYRMCIVP--KAAEKASGAEGLEN-MKVVGCRNLKDV 210
IGL GE+R V + + R+ AKLG+ +VP +K GA G + + + +L+DV
Sbjct: 315 IGLSGEVRAVAQADTRLKEAAKLGFDQALVPARPRKDKGGGAAGAASPLAIRSIGHLQDV 374
Query: 209 I 207
+
Sbjct: 375 L 375
>ref|NP_710128.1| probable ATP-dependent protease [Shigella flexneri 2a str. 301]
gi|24054956|gb|AAN45835.1|AE015447_8 probable
ATP-dependent protease [Shigella flexneri 2a str. 301]
Length = 476
Score = 89.7 bits (221), Expect = 2e-17
Identities = 45/119 (37%), Positives = 73/119 (60%)
Frame = -2
Query: 560 MIISVLIKQAGLRLQEHAVFLNVVSGLTLAETAGDLAVAAAICSSCLELPIPNDIAFIGE 381
++++VL + GL++ + VF+NVV G+ + ET+ DLA+ A+ SS + P+P D+ GE
Sbjct: 356 ILLAVLHRHGGLQMADQDVFVNVVGGVKVTETSADLALLLAMVSSLRDRPLPQDLVVFGE 415
Query: 380 IGLGGELRMVPRMEKRMYTVAKLGYRMCIVPKAAEKASGAEGLENMKVVGCRNLKDVIN 204
+GL GE+R VP ++R+ AK G+R IVP A EG M++ G + L D ++
Sbjct: 416 VGLAGEIRPVPSGQERISEAAKHGFRRAIVPAANVPKKAPEG---MQIFGVKKLSDALS 471
>ref|NP_757320.1| DNA repair protein radA [Escherichia coli CFT073]
gi|26111713|gb|AAN83894.1|AE016772_72 DNA repair protein
radA [Escherichia coli CFT073]
Length = 476
Score = 89.7 bits (221), Expect = 2e-17
Identities = 45/119 (37%), Positives = 73/119 (60%)
Frame = -2
Query: 560 MIISVLIKQAGLRLQEHAVFLNVVSGLTLAETAGDLAVAAAICSSCLELPIPNDIAFIGE 381
++++VL + GL++ + VF+NVV G+ + ET+ DLA+ A+ SS + P+P D+ GE
Sbjct: 356 ILLAVLHRHGGLQMADQDVFVNVVGGVKVTETSADLALLLAMVSSLRDRPLPQDLVVFGE 415
Query: 380 IGLGGELRMVPRMEKRMYTVAKLGYRMCIVPKAAEKASGAEGLENMKVVGCRNLKDVIN 204
+GL GE+R VP ++R+ AK G+R IVP A EG M++ G + L D ++
Sbjct: 416 VGLAGEIRPVPSGQERISEAAKHGFRRAIVPAANVPKKAPEG---MQIFGVKKLSDALS 471
>ref|NP_418806.1| probable ATP-dependent protease [Escherichia coli K12]
gi|134579|sp|P24554|RADA_ECOLI DNA repair protein radA
(DNA repair protein sms) gi|321880|pir||JC1417 DNA
repair protein sms - Escherichia coli (strain K-12)
gi|537229|gb|AAA97285.1| sms [Escherichia coli]
gi|581233|emb|CAA44856.1| sms [Escherichia coli]
gi|1401238|gb|AAC44380.1| RadA [Escherichia coli]
gi|1790850|gb|AAC77342.1| probable ATP-dependent
protease [Escherichia coli K12]
Length = 460
Score = 89.7 bits (221), Expect = 2e-17
Identities = 45/119 (37%), Positives = 73/119 (60%)
Frame = -2
Query: 560 MIISVLIKQAGLRLQEHAVFLNVVSGLTLAETAGDLAVAAAICSSCLELPIPNDIAFIGE 381
++++VL + GL++ + VF+NVV G+ + ET+ DLA+ A+ SS + P+P D+ GE
Sbjct: 340 ILLAVLHRHGGLQMADQDVFVNVVGGVKVTETSADLALLLAMVSSLRDRPLPQDLVVFGE 399
Query: 380 IGLGGELRMVPRMEKRMYTVAKLGYRMCIVPKAAEKASGAEGLENMKVVGCRNLKDVIN 204
+GL GE+R VP ++R+ AK G+R IVP A EG M++ G + L D ++
Sbjct: 400 VGLAGEIRPVPSGQERISEAAKHGFRRAIVPAANVPKKAPEG---MQIFGVKKLSDALS 455
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 457,433,857
Number of Sequences: 1393205
Number of extensions: 9572063
Number of successful extensions: 25801
Number of sequences better than 10.0: 272
Number of HSP's better than 10.0 without gapping: 24939
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25771
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)