Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004816A_C01 KMC004816A_c01
(756 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
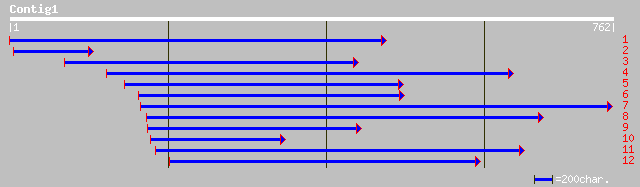
ref|NP_565951.1| expressed protein; protein id: At2g41620.1, sup... 172 5e-42
ref|NP_191294.1| putative protein; protein id: At3g57350.1 [Arab... 162 4e-39
pir||T00818 hypothetical protein At2g41620 [imported] - Arabidop... 138 1e-31
gb|EAA21911.1| hypothetical protein [Plasmodium yoelii yoelii] 34 2.6
gb|AAO50779.1| similar to Mus musculus (Mouse). Sex-determining ... 33 4.4
>ref|NP_565951.1| expressed protein; protein id: At2g41620.1, supported by cDNA:
gi_14334431, supported by cDNA: gi_21281025 [Arabidopsis
thaliana] gi|21542465|sp|O22224|Y240_ARATH Protein
At2g41620 gi|14334432|gb|AAK59414.1| unknown protein
[Arabidopsis thaliana] gi|20196948|gb|AAB84345.2|
expressed protein [Arabidopsis thaliana]
gi|21281026|gb|AAM44953.1| unknown protein [Arabidopsis
thaliana]
Length = 861
Score = 172 bits (436), Expect = 5e-42
Identities = 80/114 (70%), Positives = 98/114 (85%)
Frame = -2
Query: 755 VFEQQAVLRELESILSIHKLSRLGHHVDALREVAKLPFLPLDPRGPDVAIDVFENLSPHV 576
V EQ+ +LRELE+ILSIHKL RLG+H+DALRE+AKLPFL LDPR PD DVF++ SP+
Sbjct: 746 VMEQETILRELEAILSIHKLGRLGNHLDALREIAKLPFLHLDPRMPDATADVFQSASPYF 805
Query: 575 QACIPDLLKVALTCLDYVTDSDGSLRALRAKIASFIANNLKRNWPRDLYERVAQ 414
Q C+PDLLKVALTCLD V D+DGS+RA+R+KIA F+A+N RNWPRDLYE+VA+
Sbjct: 806 QTCVPDLLKVALTCLDNVPDTDGSIRAMRSKIAGFLASNTHRNWPRDLYEKVAR 859
>ref|NP_191294.1| putative protein; protein id: At3g57350.1 [Arabidopsis thaliana]
gi|11280530|pir||T45813 hypothetical protein F28O9.200 -
Arabidopsis thaliana gi|6735314|emb|CAB68141.1| putative
protein [Arabidopsis thaliana]
Length = 875
Score = 162 bits (411), Expect = 4e-39
Identities = 75/114 (65%), Positives = 94/114 (81%)
Frame = -2
Query: 755 VFEQQAVLRELESILSIHKLSRLGHHVDALREVAKLPFLPLDPRGPDVAIDVFENLSPHV 576
V EQ+ +LRELE+ILSIHKL+RL H+DA+REVAKLPFL LDPR PD D F+ S +
Sbjct: 760 VMEQETILRELEAILSIHKLARLNKHLDAIREVAKLPFLHLDPRQPDTTSDEFQKASSYF 819
Query: 575 QACIPDLLKVALTCLDYVTDSDGSLRALRAKIASFIANNLKRNWPRDLYERVAQ 414
Q C+PDLLKVALTCLD V D+DGS+R +R+KIA F+A+N +RNWPRDLYE++A+
Sbjct: 820 QTCVPDLLKVALTCLDNVADTDGSIRGMRSKIAGFLASNTQRNWPRDLYEKIAR 873
>pir||T00818 hypothetical protein At2g41620 [imported] - Arabidopsis thaliana
Length = 825
Score = 138 bits (347), Expect = 1e-31
Identities = 64/92 (69%), Positives = 79/92 (85%)
Frame = -2
Query: 755 VFEQQAVLRELESILSIHKLSRLGHHVDALREVAKLPFLPLDPRGPDVAIDVFENLSPHV 576
V EQ+ +LRELE+ILSIHKL RLG+H+DALRE+AKLPFL LDPR PD DVF++ SP+
Sbjct: 732 VMEQETILRELEAILSIHKLGRLGNHLDALREIAKLPFLHLDPRMPDATADVFQSASPYF 791
Query: 575 QACIPDLLKVALTCLDYVTDSDGSLRALRAKI 480
Q C+PDLLKVALTCLD V D+DGS+RA+R+K+
Sbjct: 792 QTCVPDLLKVALTCLDNVPDTDGSIRAMRSKV 823
>gb|EAA21911.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 624
Score = 33.9 bits (76), Expect = 2.6
Identities = 27/103 (26%), Positives = 45/103 (43%), Gaps = 13/103 (12%)
Frame = +1
Query: 145 VFLIILGSLL*FLNIFYKVN----------YYKATCSVYKRLAKTEEKVITKFITREFLL 294
+ II+ +L + N F+K N YY + +YK ++ I +E +
Sbjct: 488 IICIIIKTLTPYNNNFFKENTKEYNHILFIYYDISYLIYKNYILQNDEKIKNNNLKELQI 547
Query: 295 QKDQ*VLYKFYTENLHLNILLPSLIQRQQH*PSYF---HFYNL 414
QKD Y ++T+N+ I L +L ++ F HFY L
Sbjct: 548 QKDAMSSYIYFTDNMLYKIKLFNLFNFEKKIVENFIHTHFYTL 590
>gb|AAO50779.1| similar to Mus musculus (Mouse). Sex-determining region Y protein
(Testis-determining factor) [Dictyostelium discoideum]
Length = 1461
Score = 33.1 bits (74), Expect = 4.4
Identities = 19/62 (30%), Positives = 28/62 (44%), Gaps = 5/62 (8%)
Frame = +2
Query: 509 HQSP-LHSPDK*EQPSKDPEYKLGHEEINSQTH----QWQHQGPGDLEGEMEVLQPPLMH 673
HQ P H + +QP + P ++ H + N Q H Q QH L+ + + Q H
Sbjct: 947 HQQPHQHQQQQHQQPHQQPHHQQQHHQNNHQQHQQHQQHQHHQQQQLQQQQQQQQQQQHH 1006
Query: 674 QH 679
QH
Sbjct: 1007 QH 1008
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 589,105,357
Number of Sequences: 1393205
Number of extensions: 12222486
Number of successful extensions: 26968
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 25808
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26946
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36877108757
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)