Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004810A_C01 KMC004810A_c01
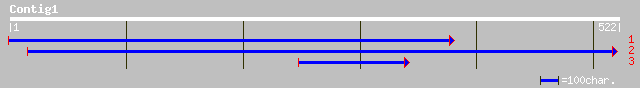
(522 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P19375|H1E_STRPU HISTONE H1, EARLY EMBRYONIC gi|10248|emb|CAA... 43 0.002
ref|NP_033766.1| AE binding protein 1; AE-binding protein 1 [Mus... 40 0.016
gb|AAC47461.1| thrombospondin-related anonymous protein [Plasmod... 39 0.048
gb|AAA63544.1| cytochrome c oxidase III 38 0.081
gb|AAA32122.1| cytochrome c oxidase III gi|447158|prf||1913438A ... 38 0.081
>sp|P19375|H1E_STRPU HISTONE H1, EARLY EMBRYONIC gi|10248|emb|CAA24644.1| histone early
H1 [Strongylocentrotus purpuratus]
Length = 205
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/55 (36%), Positives = 29/55 (52%)
Frame = +3
Query: 339 KPAKNTCPAKRERKERYRNQTKEQKTKTPKETPKKPHSKTRRRNKTPSRKQANSP 503
+P K K+ +K + K++K KTPK+ PKKP +K KTP + A P
Sbjct: 134 RPKKVKAAPKKAKKPVKKTTEKKEKKKTPKKAPKKPAAKKSTPKKTPKKAAAKKP 188
Score = 31.6 bits (70), Expect = 5.8
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Frame = +3
Query: 339 KPAKNTCPAKRERKERYRNQTKEQ-KTKTPKETPKKPHSKTRR--RNKTPSRKQA 494
KP K T K ++K + K K TPK+TPKK +K + + K P+ K+A
Sbjct: 147 KPVKKTTEKKEKKKTPKKAPKKPAAKKSTPKKTPKKAAAKKPKTAKPKKPAXKKA 201
>ref|NP_033766.1| AE binding protein 1; AE-binding protein 1 [Mus musculus]
gi|3288914|gb|AAC25584.1| aortic carboxypeptidase-like
protein ACLP [Mus musculus]
Length = 1128
Score = 40.0 bits (92), Expect = 0.016
Identities = 28/83 (33%), Positives = 40/83 (47%)
Frame = +3
Query: 264 REQRGYIMNQNMQTIPLKGGR*RT*KPAKNTCPAKRERKERYRNQTKEQKTKTPKETPKK 443
+E++G T PL+G T KP + A ++ KE+ TK+ K K PK T KK
Sbjct: 91 KEKKGKKDKGPKATKPLEGSTRPTKKPKEKPPKATKKPKEKPPKATKKPKEKPPKAT-KK 149
Query: 444 PHSKTRRRNKTPSRKQANSPLNP 512
P K + K PS + S + P
Sbjct: 150 PKEKPPKATKRPSAGKKFSTVAP 172
>gb|AAC47461.1| thrombospondin-related anonymous protein [Plasmodium gallinaceum]
Length = 614
Score = 38.5 bits (88), Expect = 0.048
Identities = 17/61 (27%), Positives = 34/61 (54%)
Frame = +1
Query: 322 EDKEPESLPKTPAPRKENEKRDIGTKPRNKRPKRQRKPQKNHTAKQEGETKPLAENRPIL 501
E+K+PES+P+ P E+++ + P K P+ + +K + +E + +P+ E + I
Sbjct: 334 EEKKPESVPEEKEPESVPEEKEPESVPEEKEPESAPEEKKPESDPEEKKLEPIPEGKKIE 393
Query: 502 P 504
P
Sbjct: 394 P 394
Score = 36.6 bits (83), Expect = 0.18
Identities = 16/61 (26%), Positives = 32/61 (52%)
Frame = +1
Query: 322 EDKEPESLPKTPAPRKENEKRDIGTKPRNKRPKRQRKPQKNHTAKQEGETKPLAENRPIL 501
E+KEPES+P+ P E+++ + P K+P+ + +K + + +P+ E +
Sbjct: 343 EEKEPESVPEEKEPESVPEEKEPESAPEEKKPESDPEEKKLEPIPEGKKIEPIPEEEKLE 402
Query: 502 P 504
P
Sbjct: 403 P 403
Score = 32.0 bits (71), Expect = 4.5
Identities = 16/61 (26%), Positives = 31/61 (50%)
Frame = +1
Query: 322 EDKEPESLPKTPAPRKENEKRDIGTKPRNKRPKRQRKPQKNHTAKQEGETKPLAENRPIL 501
E+KEPES P+ P + E++ + P K+ + + +K +E + + + E+R
Sbjct: 361 EEKEPESAPEEKKPESDPEEKKLEPIPEGKKIEPIPEEEKLEPIPEEKKPESVTEDRESE 420
Query: 502 P 504
P
Sbjct: 421 P 421
Score = 31.6 bits (70), Expect = 5.8
Identities = 13/57 (22%), Positives = 31/57 (53%)
Frame = +1
Query: 322 EDKEPESLPKTPAPRKENEKRDIGTKPRNKRPKRQRKPQKNHTAKQEGETKPLAENR 492
E+K+PE +P+ P E+++ + P K+P+ + ++ + +E E + + E +
Sbjct: 307 EEKKPEPVPEEKKPESAPEEKNPESVPEEKKPESVPEEKEPESVPEEKEPESVPEEK 363
Score = 31.6 bits (70), Expect = 5.8
Identities = 15/57 (26%), Positives = 30/57 (52%)
Frame = +1
Query: 322 EDKEPESLPKTPAPRKENEKRDIGTKPRNKRPKRQRKPQKNHTAKQEGETKPLAENR 492
E+KEPES+P+ P E++ + P K+ + + +K +E + +P+ E +
Sbjct: 352 EEKEPESVPEEKEPESAPEEKKPESDPEEKKLEPIPEGKKIEPIPEEEKLEPIPEEK 408
Score = 31.2 bits (69), Expect = 7.6
Identities = 14/56 (25%), Positives = 28/56 (50%)
Frame = +1
Query: 325 DKEPESLPKTPAPRKENEKRDIGTKPRNKRPKRQRKPQKNHTAKQEGETKPLAENR 492
D+EPE +P+ P E++ + P K P+ + +K + +E E + + E +
Sbjct: 299 DEEPEPIPEEKKPEPVPEEKKPESAPEEKNPESVPEEKKPESVPEEKEPESVPEEK 354
>gb|AAA63544.1| cytochrome c oxidase III
Length = 111
Score = 37.7 bits (86), Expect = 0.081
Identities = 22/57 (38%), Positives = 30/57 (52%), Gaps = 8/57 (14%)
Frame = -1
Query: 474 FCFSFLFCCVVF-LGFPLAFWSFVPWFGSYISLFVLFSRG-------RCFWQAFRFF 328
FC ++FCC++ GF + +W F+ F I+ F LF G CFW FRFF
Sbjct: 51 FCLPYMFCCLLCDYGF-VFYWYFLDLFNLLINTFYLFVSGLFVNFFVLCFW--FRFF 104
>gb|AAA32122.1| cytochrome c oxidase III gi|447158|prf||1913438A cytochrome c
oxidase
Length = 288
Score = 37.7 bits (86), Expect = 0.081
Identities = 22/57 (38%), Positives = 30/57 (52%), Gaps = 8/57 (14%)
Frame = -1
Query: 474 FCFSFLFCCVVF-LGFPLAFWSFVPWFGSYISLFVLFSRG-------RCFWQAFRFF 328
FC ++FCC++ GF + +W F+ F I+ F LF G CFW FRFF
Sbjct: 123 FCLPYMFCCLLCDYGF-VFYWYFLDLFNLLINTFYLFVSGLFVNFFVLCFW--FRFF 176
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 445,110,809
Number of Sequences: 1393205
Number of extensions: 9929064
Number of successful extensions: 57283
Number of sequences better than 10.0: 353
Number of HSP's better than 10.0 without gapping: 46526
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 55328
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)