Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
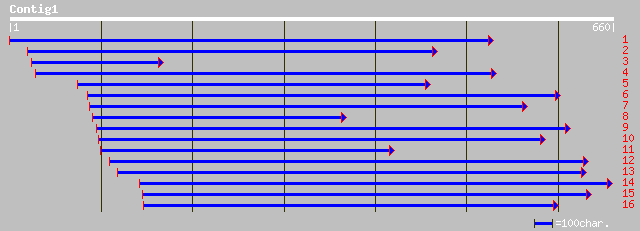
Query= KMC004809A_C01 KMC004809A_c01
(641 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_201247.1| putative protein; protein id: At5g64420.1 [Arab... 54 2e-06
gb|EAA17262.1| CCAAT-box DNA binding protein subunit B [Plasmodi... 34 1.4
gb|AAN46932.1|AF472533_37 unknown [Campylobacter jejuni] 33 2.5
ref|XP_114418.2| similar to KIAA1729 protein [Homo sapiens] 33 4.2
dbj|BAB21820.1| KIAA1729 protein [Homo sapiens] 33 4.2
>ref|NP_201247.1| putative protein; protein id: At5g64420.1 [Arabidopsis thaliana]
Length = 1306
Score = 53.5 bits (127), Expect = 2e-06
Identities = 25/59 (42%), Positives = 34/59 (57%)
Frame = -3
Query: 636 NMPSKSARRSEVNKFCVKCFEILSKLNLTKPLVKALPPDAQAALEAQLGDKFTRLKKLD 460
NMP RR++V KFC + F+++S L LTK +K L D + A E GD F LK +
Sbjct: 1247 NMPEAKVRRAQVRKFCGRIFQMVSSLKLTKSFLKGLGQDGRTACEDAFGDLFLNLKNTE 1305
>gb|EAA17262.1| CCAAT-box DNA binding protein subunit B [Plasmodium yoelii yoelii]
Length = 850
Score = 34.3 bits (77), Expect = 1.4
Identities = 26/78 (33%), Positives = 39/78 (49%)
Frame = +1
Query: 133 ILFTQSKSHNMQKKGSQNNT*PETNKINNFIVYQNSTPILLLKALKLQSIMKDQITKMFK 312
ILFT++ S N + K QNN NN ++ +N+ I L +KL + + T
Sbjct: 32 ILFTENISINSRNKNGQNN----YEHRNNIVLAKNTIDIYKLWLVKLVTYEDNVET---S 84
Query: 313 DQITAFGHNTVFRN*LNE 366
+ IT NT+F N +NE
Sbjct: 85 NIITTNNKNTIFTNSINE 102
>gb|AAN46932.1|AF472533_37 unknown [Campylobacter jejuni]
Length = 136
Score = 33.5 bits (75), Expect = 2.5
Identities = 14/35 (40%), Positives = 23/35 (65%)
Frame = -2
Query: 124 IFISIFFSSANFDYRYFRPNVYLKFNIILVFFYMM 20
+F+ ++S+ANF YFR L+F IIL+ F ++
Sbjct: 88 LFLFFYYSNANFSLNYFRKLYDLQFPIILILFIVV 122
>ref|XP_114418.2| similar to KIAA1729 protein [Homo sapiens]
Length = 1074
Score = 32.7 bits (73), Expect = 4.2
Identities = 18/65 (27%), Positives = 31/65 (47%)
Frame = +1
Query: 109 KWI*KFKFILFTQSKSHNMQKKGSQNNT*PETNKINNFIVYQNSTPILLLKALKLQSIMK 288
KW+ + FI+ S HN K + T + K NN ++ + P + LK L S+ K
Sbjct: 433 KWVRSYDFIM-PNSSVHNNGKSFINSETIEDFQKKNNLYPHRTAFPSVALKGHSLASVFK 491
Query: 289 DQITK 303
+ + +
Sbjct: 492 NSVLR 496
>dbj|BAB21820.1| KIAA1729 protein [Homo sapiens]
Length = 1020
Score = 32.7 bits (73), Expect = 4.2
Identities = 18/65 (27%), Positives = 31/65 (47%)
Frame = +1
Query: 109 KWI*KFKFILFTQSKSHNMQKKGSQNNT*PETNKINNFIVYQNSTPILLLKALKLQSIMK 288
KW+ + FI+ S HN K + T + K NN ++ + P + LK L S+ K
Sbjct: 379 KWVRSYDFIM-PNSSVHNNGKSFINSETIEDFQKKNNLYPHRTAFPSVALKGHSLASVFK 437
Query: 289 DQITK 303
+ + +
Sbjct: 438 NSVLR 442
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,450,453
Number of Sequences: 1393205
Number of extensions: 9932997
Number of successful extensions: 22588
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 21909
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22581
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)