Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004781A_C01 KMC004781A_c01
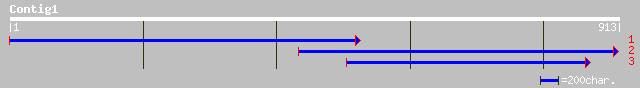
(913 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||H86202 hypothetical protein [imported] - Arabidopsis thalia... 115 1e-24
pir||G84709 probable lipase [imported] - Arabidopsis thaliana 107 2e-22
ref|NP_565701.1| putative lipase; protein id: At2g30550.1, suppo... 107 2e-22
ref|NP_564590.1| expressed protein; protein id: At1g51440.1, sup... 100 4e-20
dbj|BAB89206.1| lipase-like protein [Oryza sativa (japonica cult... 66 8e-10
>pir||H86202 hypothetical protein [imported] - Arabidopsis thaliana
gi|7523699|gb|AAF63138.1|AC011001_8 Similar to lipases
[Arabidopsis thaliana] gi|20466266|gb|AAM20450.1|
lipase, putative [Arabidopsis thaliana]
Length = 515
Score = 115 bits (287), Expect = 1e-24
Identities = 53/81 (65%), Positives = 63/81 (77%), Gaps = 7/81 (8%)
Frame = +1
Query: 655 YHGKGQRFILSTGRDPALVNKACDCLKDHYLVPPHWRQDENKGMIRGNDGRWVQPERPKL 834
YHGKGQRF+LS+GRDPALVNKA D LKDH++VPP+WRQD NKGM+R DGRW+QP+R +
Sbjct: 434 YHGKGQRFVLSSGRDPALVNKASDFLKDHFMVPPYWRQDANKGMVRNTDGRWIQPDRIRA 493
Query: 835 DD-HPEDM------LHHLRQL 876
DD H D+ LHH QL
Sbjct: 494 DDQHAPDIHQLLTQLHHPSQL 514
>pir||G84709 probable lipase [imported] - Arabidopsis thaliana
Length = 447
Score = 107 bits (267), Expect = 2e-22
Identities = 48/77 (62%), Positives = 60/77 (77%), Gaps = 1/77 (1%)
Frame = +1
Query: 655 YHGKGQRFILSTGRDPALVNKACDCLKDHYLVPPHWRQDENKGMIRGNDGRWVQPERPKL 834
YHGKG+RF+LS+GRD ALVNKA D LK+H +PP WRQD NKGM+R ++GRW+Q ER +
Sbjct: 367 YHGKGERFVLSSGRDHALVNKASDFLKEHLQIPPFWRQDANKGMVRNSEGRWIQAERLRF 426
Query: 835 DD-HPEDMLHHLRQLGL 882
+D H D+ HHL QL L
Sbjct: 427 EDHHSPDIHHHLSQLRL 443
>ref|NP_565701.1| putative lipase; protein id: At2g30550.1, supported by cDNA:
207043., supported by cDNA: gi_20268777 [Arabidopsis
thaliana] gi|20198327|gb|AAB63082.2| putative lipase
[Arabidopsis thaliana] gi|20268778|gb|AAM14092.1|
putative lipase [Arabidopsis thaliana]
gi|21554152|gb|AAM63231.1| putative lipase [Arabidopsis
thaliana] gi|23296757|gb|AAN13163.1| putative lipase
[Arabidopsis thaliana]
Length = 529
Score = 107 bits (267), Expect = 2e-22
Identities = 48/77 (62%), Positives = 60/77 (77%), Gaps = 1/77 (1%)
Frame = +1
Query: 655 YHGKGQRFILSTGRDPALVNKACDCLKDHYLVPPHWRQDENKGMIRGNDGRWVQPERPKL 834
YHGKG+RF+LS+GRD ALVNKA D LK+H +PP WRQD NKGM+R ++GRW+Q ER +
Sbjct: 449 YHGKGERFVLSSGRDHALVNKASDFLKEHLQIPPFWRQDANKGMVRNSEGRWIQAERLRF 508
Query: 835 DD-HPEDMLHHLRQLGL 882
+D H D+ HHL QL L
Sbjct: 509 EDHHSPDIHHHLSQLRL 525
>ref|NP_564590.1| expressed protein; protein id: At1g51440.1, supported by cDNA:
gi_15983395 [Arabidopsis thaliana]
gi|25405467|pir||F96552 hypothetical protein F5D21.19
[imported] - Arabidopsis thaliana
gi|12325376|gb|AAG52635.1|AC024261_22 hypothetical
protein; 69776-68193 [Arabidopsis thaliana]
gi|15983396|gb|AAL11566.1|AF424572_1 At1g51440/F5D21_19
[Arabidopsis thaliana] gi|22655390|gb|AAM98287.1|
At1g51440/F5D21_19 [Arabidopsis thaliana]
Length = 527
Score = 100 bits (248), Expect = 4e-20
Identities = 50/86 (58%), Positives = 61/86 (70%), Gaps = 6/86 (6%)
Frame = +1
Query: 655 YHGKGQ----RFILSTGRDPALVNKACDCLKDHYLVPPHWRQDENKGMIRGNDGRWVQPE 822
YHGK + RF L T RD ALVNK+CD L+ Y VPP WRQDENKGM++ DG+WV P+
Sbjct: 435 YHGKDEEAEKRFCLVTKRDIALVNKSCDFLRGEYHVPPCWRQDENKGMVKNGDGQWVLPD 494
Query: 823 RPKLDDH-PEDMLHHLRQ-LGLTSSD 894
RP L+ H PED+ HHL+Q LG + D
Sbjct: 495 RPLLEPHGPEDIAHHLQQVLGKVNDD 520
>dbj|BAB89206.1| lipase-like protein [Oryza sativa (japonica cultivar-group)]
Length = 403
Score = 65.9 bits (159), Expect = 8e-10
Identities = 33/67 (49%), Positives = 39/67 (57%), Gaps = 1/67 (1%)
Frame = +1
Query: 655 YHGKGQRFILSTGRDPALVNKACDCLKDHYLVPPHWRQDENKGMIRGNDGRWV-QPERPK 831
+HG + F L RD ALVNK DCL D Y VP W+ NK M++G DGRWV Q P
Sbjct: 335 WHGDHRGFELVVDRDVALVNKFDDCLADEYPVPVRWKVHHNKSMVKGPDGRWVLQDHEPD 394
Query: 832 LDDHPED 852
DD +D
Sbjct: 395 DDDDDDD 401
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 728,110,906
Number of Sequences: 1393205
Number of extensions: 15755937
Number of successful extensions: 32748
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 31000
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32725
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49918505760
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)