Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
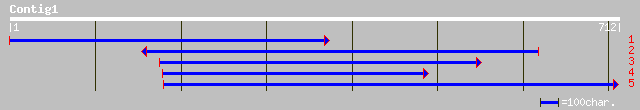
Query= KMC004759A_C01 KMC004759A_c01
(711 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN41283.1| putative scarecrow protein [Arabidopsis thaliana] 226 2e-58
pir||T51236 scarecrow-like protein 5 [imported] - Arabidopsis th... 226 2e-58
ref|NP_175475.1| scarecrow-like protein; protein id: At1g50600.1... 226 2e-58
gb|AAK59436.2| putative scarecrow protein [Arabidopsis thaliana] 226 2e-58
gb|AAK62666.1| F17J6.12/F17J6.12 [Arabidopsis thaliana] 225 5e-58
>gb|AAN41283.1| putative scarecrow protein [Arabidopsis thaliana]
Length = 597
Score = 226 bits (577), Expect = 2e-58
Identities = 110/142 (77%), Positives = 119/142 (83%)
Frame = -3
Query: 706 KCLSPKVVTLVEQEFNTNNAPFLQRFVETMNYYQAVFESIDVVLPREHKERINVEQHCLA 527
K LSP VVTLVEQE NTN APFL RFVETMN+Y AVFESIDV L R+HKERINVEQHCLA
Sbjct: 456 KHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLA 515
Query: 526 REVVNLVACEGAERVERHELLNKWRMRFASAGFTPYPLNSYINSSIKDLLESYRGHYTLE 347
REVVNL+ACEG ER ERHE L KWR RF AGF PYPL+SY+N++IK LLESY YTLE
Sbjct: 516 REVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLE 575
Query: 346 ERDGALFLGWMNQVLVASCAWR 281
ERDGAL+LGW NQ L+ SCAWR
Sbjct: 576 ERDGALYLGWKNQPLITSCAWR 597
>pir||T51236 scarecrow-like protein 5 [imported] - Arabidopsis thaliana
(fragment) gi|4580517|gb|AAD24405.1|AF036302_1
scarecrow-like 5 [Arabidopsis thaliana]
Length = 306
Score = 226 bits (577), Expect = 2e-58
Identities = 110/142 (77%), Positives = 119/142 (83%)
Frame = -3
Query: 706 KCLSPKVVTLVEQEFNTNNAPFLQRFVETMNYYQAVFESIDVVLPREHKERINVEQHCLA 527
K LSP VVTLVEQE NTN APFL RFVETMN+Y AVFESIDV L R+HKERINVEQHCLA
Sbjct: 165 KHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLA 224
Query: 526 REVVNLVACEGAERVERHELLNKWRMRFASAGFTPYPLNSYINSSIKDLLESYRGHYTLE 347
REVVNL+ACEG ER ERHE L KWR RF AGF PYPL+SY+N++IK LLESY YTLE
Sbjct: 225 REVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLE 284
Query: 346 ERDGALFLGWMNQVLVASCAWR 281
ERDGAL+LGW NQ L+ SCAWR
Sbjct: 285 ERDGALYLGWKNQPLITSCAWR 306
>ref|NP_175475.1| scarecrow-like protein; protein id: At1g50600.1, supported by cDNA:
gi_14334475, supported by cDNA: gi_14517551, supported
by cDNA: gi_17979104 [Arabidopsis thaliana]
gi|25405382|pir||E96542 scarecrow-like protein
[imported] - Arabidopsis thaliana
gi|9454552|gb|AAF87875.1|AC012561_8 Putative
transcription factor [Arabidopsis thaliana]
gi|12322334|gb|AAG51190.1|AC079279_11 scarecrow-like
protein [Arabidopsis thaliana]
Length = 526
Score = 226 bits (577), Expect = 2e-58
Identities = 110/142 (77%), Positives = 119/142 (83%)
Frame = -3
Query: 706 KCLSPKVVTLVEQEFNTNNAPFLQRFVETMNYYQAVFESIDVVLPREHKERINVEQHCLA 527
K LSP VVTLVEQE NTN APFL RFVETMN+Y AVFESIDV L R+HKERINVEQHCLA
Sbjct: 385 KHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLA 444
Query: 526 REVVNLVACEGAERVERHELLNKWRMRFASAGFTPYPLNSYINSSIKDLLESYRGHYTLE 347
REVVNL+ACEG ER ERHE L KWR RF AGF PYPL+SY+N++IK LLESY YTLE
Sbjct: 445 REVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLE 504
Query: 346 ERDGALFLGWMNQVLVASCAWR 281
ERDGAL+LGW NQ L+ SCAWR
Sbjct: 505 ERDGALYLGWKNQPLITSCAWR 526
>gb|AAK59436.2| putative scarecrow protein [Arabidopsis thaliana]
Length = 587
Score = 226 bits (577), Expect = 2e-58
Identities = 110/142 (77%), Positives = 119/142 (83%)
Frame = -3
Query: 706 KCLSPKVVTLVEQEFNTNNAPFLQRFVETMNYYQAVFESIDVVLPREHKERINVEQHCLA 527
K LSP VVTLVEQE NTN APFL RFVETMN+Y AVFESIDV L R+HKERINVEQHCLA
Sbjct: 446 KHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLA 505
Query: 526 REVVNLVACEGAERVERHELLNKWRMRFASAGFTPYPLNSYINSSIKDLLESYRGHYTLE 347
REVVNL+ACEG ER ERHE L KWR RF AGF PYPL+SY+N++IK LLESY YTLE
Sbjct: 506 REVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLE 565
Query: 346 ERDGALFLGWMNQVLVASCAWR 281
ERDGAL+LGW NQ L+ SCAWR
Sbjct: 566 ERDGALYLGWKNQPLITSCAWR 587
>gb|AAK62666.1| F17J6.12/F17J6.12 [Arabidopsis thaliana]
Length = 526
Score = 225 bits (573), Expect = 5e-58
Identities = 109/142 (76%), Positives = 119/142 (83%)
Frame = -3
Query: 706 KCLSPKVVTLVEQEFNTNNAPFLQRFVETMNYYQAVFESIDVVLPREHKERINVEQHCLA 527
K LSP VVTLVEQE NTN APFL RFVETMN+Y AVFESIDV L R+HKERINVEQHCLA
Sbjct: 385 KHLSPNVVTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLA 444
Query: 526 REVVNLVACEGAERVERHELLNKWRMRFASAGFTPYPLNSYINSSIKDLLESYRGHYTLE 347
REVVNL+ACEG ER ERHE L KWR RF AGF PYPL+SY+N++I+ LLESY YTLE
Sbjct: 445 REVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIEGLLESYSEKYTLE 504
Query: 346 ERDGALFLGWMNQVLVASCAWR 281
ERDGAL+LGW NQ L+ SCAWR
Sbjct: 505 ERDGALYLGWKNQPLITSCAWR 526
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 566,987,991
Number of Sequences: 1393205
Number of extensions: 11481461
Number of successful extensions: 26086
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 25281
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25941
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32654539052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)