Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004754A_C01 KMC004754A_c01
(613 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
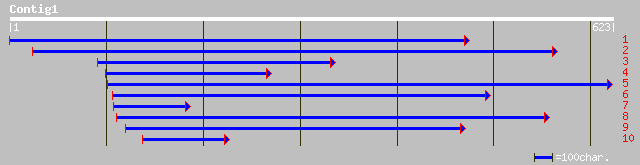
ref|NP_567456.1| Expressed protein; protein id: At4g15093.1 [Ara... 146 2e-34
dbj|BAC06208.1| P0018C10.9 [Oryza sativa (japonica cultivar-grou... 143 2e-33
dbj|BAC06209.1| P0018C10.10 [Oryza sativa (japonica cultivar-gro... 127 1e-28
pir||F71414 hypothetical protein - Arabidopsis thaliana gi|22448... 127 1e-28
ref|NP_655723.1| LigB, Catalytic LigB subunit of aromatic ring-o... 86 4e-16
>ref|NP_567456.1| Expressed protein; protein id: At4g15093.1 [Arabidopsis thaliana]
gi|15293029|gb|AAK93625.1| unknown protein [Arabidopsis
thaliana] gi|21436089|gb|AAM51245.1| unknown protein
[Arabidopsis thaliana] gi|26452573|dbj|BAC43371.1|
unknown protein [Arabidopsis thaliana]
Length = 269
Score = 146 bits (369), Expect = 2e-34
Identities = 68/112 (60%), Positives = 91/112 (80%), Gaps = 2/112 (1%)
Frame = -3
Query: 608 LPDEGVLILGSGSAVPHLRALERHATVAAP--WAVEFDNWLKEALLEGRYEDVNHYEQKA 435
L DEGVLI+GSGSA +LR L+ + T +P WA+EFD+WL+++LL+GRY DVN +E+KA
Sbjct: 158 LKDEGVLIIGSGSATHNLRKLDFNITDGSPVPWALEFDHWLRDSLLQGRYGDVNEWEEKA 217
Query: 434 PHAKKAHPWPDHFYPLHVAIGAAGENSKAKLIHSSIDLGSLSYASYQFTSDV 279
P+AK AHPWP+H YPLHV +GAAG ++KA+ IH+S LG+LSY+SY FTS +
Sbjct: 218 PNAKMAHPWPEHLYPLHVVMGAAGGDAKAEQIHTSWQLGTLSYSSYSFTSSL 269
>dbj|BAC06208.1| P0018C10.9 [Oryza sativa (japonica cultivar-group)]
gi|22202675|dbj|BAC07333.1| P0471B04.17 [Oryza sativa
(japonica cultivar-group)]
Length = 266
Score = 143 bits (361), Expect = 2e-33
Identities = 68/109 (62%), Positives = 82/109 (74%)
Frame = -3
Query: 611 PLPDEGVLILGSGSAVPHLRALERHATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAP 432
PL D+GVL+LGSGSA +LR + T WA EFD WL+EALL GR++DV YE+KAP
Sbjct: 155 PLRDDGVLVLGSGSATHNLRRMGPEGTPVPQWAAEFDGWLQEALLGGRHDDVKRYEEKAP 214
Query: 431 HAKKAHPWPDHFYPLHVAIGAAGENSKAKLIHSSIDLGSLSYASYQFTS 285
H + AHP PDHF PLHVA+GAAGE +KA+LIH S SLSYASY+FT+
Sbjct: 215 HGRVAHPSPDHFLPLHVALGAAGEGAKAELIHRSWSNASLSYASYRFTT 263
>dbj|BAC06209.1| P0018C10.10 [Oryza sativa (japonica cultivar-group)]
gi|22202676|dbj|BAC07334.1| P0471B04.18 [Oryza sativa
(japonica cultivar-group)]
Length = 280
Score = 127 bits (319), Expect = 1e-28
Identities = 66/112 (58%), Positives = 83/112 (73%), Gaps = 3/112 (2%)
Frame = -3
Query: 611 PLPDEGVLILGSGSAVPHLRALERHA--TVAAPWAVEFDNWLKEALLEG-RYEDVNHYEQ 441
PL D+GVLILGSG+A +L + A T WA EFD WL+EALL G R++DV YE+
Sbjct: 157 PLRDDGVLILGSGNATHNLSCMAPVAEGTPVPQWAAEFDGWLQEALLAGGRHDDVKQYEE 216
Query: 440 KAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKLIHSSIDLGSLSYASYQFTS 285
KAP+ K AHP PDHF PLHVA+GAAGE++KA+LIH S +LS+ASY+FT+
Sbjct: 217 KAPNGKMAHPSPDHFLPLHVALGAAGEDAKAELIHHSWYNATLSHASYRFTT 268
>pir||F71414 hypothetical protein - Arabidopsis thaliana
gi|2244866|emb|CAB10288.1| hypothetical protein
[Arabidopsis thaliana] gi|7268255|emb|CAB78551.1|
hypothetical protein [Arabidopsis thaliana]
Length = 1705
Score = 127 bits (319), Expect = 1e-28
Identities = 59/98 (60%), Positives = 79/98 (80%), Gaps = 2/98 (2%)
Frame = -3
Query: 608 LPDEGVLILGSGSAVPHLRALERHATVAAP--WAVEFDNWLKEALLEGRYEDVNHYEQKA 435
L DEGVLI+GSGSA +LR L+ + T +P WA+EFD+WL+++LL+GRY DVN +E+KA
Sbjct: 936 LKDEGVLIIGSGSATHNLRKLDFNITDGSPVPWALEFDHWLRDSLLQGRYGDVNEWEEKA 995
Query: 434 PHAKKAHPWPDHFYPLHVAIGAAGENSKAKLIHSSIDL 321
P+AK AHPWP+H YPLHV +GAAG ++KA+ IH+S L
Sbjct: 996 PNAKMAHPWPEHLYPLHVVMGAAGGDAKAEQIHTSWQL 1033
>ref|NP_655723.1| LigB, Catalytic LigB subunit of aromatic ring-opening dioxygenase
[Bacillus anthracis A2012]
Length = 253
Score = 85.9 bits (211), Expect = 4e-16
Identities = 44/106 (41%), Positives = 70/106 (65%)
Frame = -3
Query: 608 LPDEGVLILGSGSAVPHLRALERHATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPH 429
L E +L++GSG V +LRAL+ + T WA+EFD+W+ + + + + ++E+ APH
Sbjct: 150 LGQEDILVIGSGVTVHNLRALKWNQTTPEQWAIEFDDWIIKHMQAADKDALFNWEKNAPH 209
Query: 428 AKKAHPWPDHFYPLHVAIGAAGENSKAKLIHSSIDLGSLSYASYQF 291
A+ A P +HF PL +A+G +GEN+ ++IH S +LG+LSY QF
Sbjct: 210 AQLAVPRAEHFVPLFIAMG-SGENN-GEVIHRSYELGTLSYLCLQF 253
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 510,924,147
Number of Sequences: 1393205
Number of extensions: 10603923
Number of successful extensions: 25535
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 24760
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25493
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)