Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004750A_C01 KMC004750A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
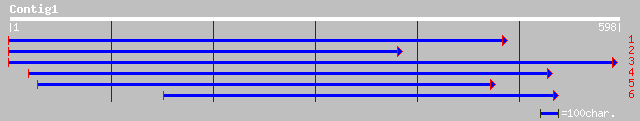
gb|AAD28760.1|AF130253_1 membrane related protein CP5 [Arabidops... 83 3e-15
gb|AAM91533.1| membrane related protein CP5, putative [Arabidops... 83 3e-15
ref|NP_176653.1| membrane related protein CP5, putative; protein... 83 3e-15
gb|AAL57201.1|AF355597_1 putative nodule membrane protein [Medic... 70 2e-11
dbj|BAB11579.1| membrane related protein-like [Arabidopsis thali... 39 0.037
>gb|AAD28760.1|AF130253_1 membrane related protein CP5 [Arabidopsis thaliana]
Length = 387
Score = 82.8 bits (203), Expect = 3e-15
Identities = 38/47 (80%), Positives = 46/47 (97%)
Frame = -3
Query: 554 KPIGRNIPKLLVVGGAIALACTLDQGLVTKAVIFGVARRFAKIGRRL 414
KP G+NIPK+LVVGGAIALACTLD+GL+TKAVIFGVARRFA++G+R+
Sbjct: 341 KPTGKNIPKILVVGGAIALACTLDKGLLTKAVIFGVARRFARMGKRM 387
>gb|AAM91533.1| membrane related protein CP5, putative [Arabidopsis thaliana]
gi|23197608|gb|AAN15331.1| membrane related protein CP5,
putative [Arabidopsis thaliana]
Length = 385
Score = 82.8 bits (203), Expect = 3e-15
Identities = 38/47 (80%), Positives = 46/47 (97%)
Frame = -3
Query: 554 KPIGRNIPKLLVVGGAIALACTLDQGLVTKAVIFGVARRFAKIGRRL 414
KP G+NIPK+LVVGGAIALACTLD+GL+TKAVIFGVARRFA++G+R+
Sbjct: 339 KPTGKNIPKILVVGGAIALACTLDKGLLTKAVIFGVARRFARMGKRM 385
>ref|NP_176653.1| membrane related protein CP5, putative; protein id: At1g64720.1,
supported by cDNA: gi_4741928 [Arabidopsis thaliana]
gi|25343851|pir||D96670 probable membrane related
protein F13O11.4 [imported] - Arabidopsis thaliana
gi|5042409|gb|AAD38248.1|AC006193_4 Putative membrane
related protein [Arabidopsis thaliana]
Length = 385
Score = 82.8 bits (203), Expect = 3e-15
Identities = 38/47 (80%), Positives = 46/47 (97%)
Frame = -3
Query: 554 KPIGRNIPKLLVVGGAIALACTLDQGLVTKAVIFGVARRFAKIGRRL 414
KP G+NIPK+LVVGGAIALACTLD+GL+TKAVIFGVARRFA++G+R+
Sbjct: 339 KPTGKNIPKILVVGGAIALACTLDKGLLTKAVIFGVARRFARMGKRM 385
>gb|AAL57201.1|AF355597_1 putative nodule membrane protein [Medicago sativa]
Length = 424
Score = 70.1 bits (170), Expect = 2e-11
Identities = 34/51 (66%), Positives = 43/51 (83%), Gaps = 2/51 (3%)
Frame = -3
Query: 563 SSVKPIGRNIPKLLVVGGAIALACTLDQGLVTKAVIFGVARR--FAKIGRR 417
S KP G +PK+LV+GGA+ALAC+LD+GLVTKAVIFGVA+R FA +G+R
Sbjct: 374 SDDKPKGMTVPKMLVIGGAVALACSLDKGLVTKAVIFGVAKRFGFANLGKR 424
>dbj|BAB11579.1| membrane related protein-like [Arabidopsis thaliana]
Length = 400
Score = 39.3 bits (90), Expect = 0.037
Identities = 18/42 (42%), Positives = 27/42 (63%), Gaps = 1/42 (2%)
Frame = -3
Query: 539 NIPKLLVVGGAIALACTLDQG-LVTKAVIFGVARRFAKIGRR 417
N+ KLL++GGA+A+ C+L G V A + G +RF GR+
Sbjct: 343 NLKKLLIIGGAVAVVCSLSGGAFVPPAFLLGFGKRFVNGGRK 384
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 482,926,235
Number of Sequences: 1393205
Number of extensions: 9784095
Number of successful extensions: 19253
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 18823
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19246
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)