Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004740A_C01 KMC004740A_c01
(697 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
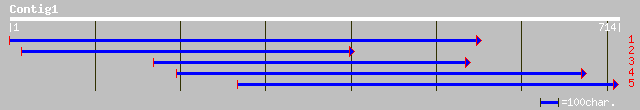
ref|NP_566004.1| VirF-interacting protein FIP1; protein id: At2g... 65 4e-19
pir||T00677 hypothetical protein At2g43970 [imported] - Arabidop... 65 4e-19
gb|AAL06505.1|AF412052_1 At2g43970/F6E13.10 [Arabidopsis thaliana] 65 4e-19
pir||T12631 DRS12 protein - common sunflower (fragment) gi|23264... 55 3e-07
ref|NP_568660.1| RRM-containing protein; protein id: At5g46250.1... 47 4e-06
>ref|NP_566004.1| VirF-interacting protein FIP1; protein id: At2g43970.1, supported
by cDNA: gi_12744918, supported by cDNA: gi_14030638,
supported by cDNA: gi_14194142 [Arabidopsis thaliana]
gi|12744919|gb|AAK06847.1| VirF-interacting protein FIP1
[Arabidopsis thaliana]
gi|14030639|gb|AAK52994.1|AF375410_1 At2g43970/F6E13.10
[Arabidopsis thaliana]
gi|14194143|gb|AAK56266.1|AF367277_1 At2g43970/F6E13.10
[Arabidopsis thaliana] gi|15810399|gb|AAL07087.1|
unknown protein [Arabidopsis thaliana]
gi|20197078|gb|AAC23405.2| expressed protein
[Arabidopsis thaliana] gi|22136548|gb|AAM91060.1|
At2g43970/F6E13.10 [Arabidopsis thaliana]
Length = 545
Score = 65.1 bits (157), Expect(2) = 4e-19
Identities = 40/94 (42%), Positives = 55/94 (57%), Gaps = 4/94 (4%)
Frame = -1
Query: 691 QGRGQKGLDVEVGCEEGHTSVPELQADDKPLEDASIPEIQPH---EHVGEEHGYD*ESGR 521
QGRG+KG D +V EE + E Q +K +D S E H + +GE+ G + +G+
Sbjct: 391 QGRGRKGHDADVEHEEDDATTSEQQPIEKQSDDCS-GEWDTHMQEQPIGEDQGNEKAAGQ 449
Query: 520 RKGRSRGRAKGRGRVHCHQN-NRVNHLGTPSSHN 422
RKGR+RGR KGRGR HQN N+ N+ +HN
Sbjct: 450 RKGRNRGRGKGRGRGQPHQNQNQNNNHSHNQNHN 483
Score = 51.6 bits (122), Expect(2) = 4e-19
Identities = 21/23 (91%), Positives = 22/23 (95%)
Frame = -2
Query: 387 PPGPRMPDGTRGFSMGRGKPISV 319
PPGPRMPDGTRGFSMGRGKP+ V
Sbjct: 520 PPGPRMPDGTRGFSMGRGKPVMV 542
>pir||T00677 hypothetical protein At2g43970 [imported] - Arabidopsis thaliana
Length = 529
Score = 65.1 bits (157), Expect(2) = 4e-19
Identities = 40/94 (42%), Positives = 55/94 (57%), Gaps = 4/94 (4%)
Frame = -1
Query: 691 QGRGQKGLDVEVGCEEGHTSVPELQADDKPLEDASIPEIQPH---EHVGEEHGYD*ESGR 521
QGRG+KG D +V EE + E Q +K +D S E H + +GE+ G + +G+
Sbjct: 375 QGRGRKGHDADVEHEEDDATTSEQQPIEKQSDDCS-GEWDTHMQEQPIGEDQGNEKAAGQ 433
Query: 520 RKGRSRGRAKGRGRVHCHQN-NRVNHLGTPSSHN 422
RKGR+RGR KGRGR HQN N+ N+ +HN
Sbjct: 434 RKGRNRGRGKGRGRGQPHQNQNQNNNHSHNQNHN 467
Score = 51.6 bits (122), Expect(2) = 4e-19
Identities = 21/23 (91%), Positives = 22/23 (95%)
Frame = -2
Query: 387 PPGPRMPDGTRGFSMGRGKPISV 319
PPGPRMPDGTRGFSMGRGKP+ V
Sbjct: 504 PPGPRMPDGTRGFSMGRGKPVMV 526
>gb|AAL06505.1|AF412052_1 At2g43970/F6E13.10 [Arabidopsis thaliana]
Length = 329
Score = 65.1 bits (157), Expect(2) = 4e-19
Identities = 40/94 (42%), Positives = 55/94 (57%), Gaps = 4/94 (4%)
Frame = -1
Query: 691 QGRGQKGLDVEVGCEEGHTSVPELQADDKPLEDASIPEIQPH---EHVGEEHGYD*ESGR 521
QGRG+KG D +V EE + E Q +K +D S E H + +GE+ G + +G+
Sbjct: 175 QGRGRKGHDADVEHEEDDATTSEQQPIEKQSDDCS-GEWDTHMQEQPIGEDQGNEKAAGQ 233
Query: 520 RKGRSRGRAKGRGRVHCHQN-NRVNHLGTPSSHN 422
RKGR+RGR KGRGR HQN N+ N+ +HN
Sbjct: 234 RKGRNRGRGKGRGRGQPHQNQNQNNNHSHNQNHN 267
Score = 51.6 bits (122), Expect(2) = 4e-19
Identities = 21/23 (91%), Positives = 22/23 (95%)
Frame = -2
Query: 387 PPGPRMPDGTRGFSMGRGKPISV 319
PPGPRMPDGTRGFSMGRGKP+ V
Sbjct: 304 PPGPRMPDGTRGFSMGRGKPVMV 326
>pir||T12631 DRS12 protein - common sunflower (fragment)
gi|2326450|emb|CAA72655.1| DRS12 protein [Helianthus
annuus] gi|18857656|emb|CAA54462.1| putative drought
tolerance protein [Helianthus annuus x Helianthus
argophyllus]
Length = 138
Score = 55.1 bits (131), Expect(2) = 3e-07
Identities = 36/92 (39%), Positives = 53/92 (57%), Gaps = 4/92 (4%)
Frame = -1
Query: 673 GLDVEVGCEEGHTSVPELQA-DDKPLEDA-SIPEIQPHEHVG-EEHGYD*ESGRRKGRSR 503
G + + G + TS E + ++K ++D ++Q HEHV E+H D E +KGR+R
Sbjct: 15 GQEDDAGPNQDDTSTSEQHSLNEKHVDDTCQQSDVQSHEHVQVEDHPNDKEGRHKKGRNR 74
Query: 502 GRAKGRGR-VHCHQNNRVNHLGTPSSHNSILT 410
GR KGRGR + H NNR NH+ S H ++T
Sbjct: 75 GRGKGRGRPQYNHHNNRGNHM---SPHLQLIT 103
Score = 21.6 bits (44), Expect(2) = 3e-07
Identities = 9/17 (52%), Positives = 9/17 (52%)
Frame = -3
Query: 386 LLGHACQMVQEDFRWVG 336
LLG ACQ Q W G
Sbjct: 115 LLGPACQTEQGGSHWTG 131
>ref|NP_568660.1| RRM-containing protein; protein id: At5g46250.1, supported by cDNA:
gi_15215747 [Arabidopsis thaliana]
gi|15215748|gb|AAK91419.1| AT5g46250/MPL12_3
[Arabidopsis thaliana] gi|23308375|gb|AAN18157.1|
At5g46250/MPL12_3 [Arabidopsis thaliana]
Length = 422
Score = 46.6 bits (109), Expect(2) = 4e-06
Identities = 19/21 (90%), Positives = 20/21 (94%)
Frame = -2
Query: 387 PPGPRMPDGTRGFSMGRGKPI 325
PPGPRMPDGTRGF+MGRGK I
Sbjct: 390 PPGPRMPDGTRGFTMGRGKAI 410
Score = 25.8 bits (55), Expect(2) = 4e-06
Identities = 16/46 (34%), Positives = 24/46 (51%), Gaps = 2/46 (4%)
Frame = -1
Query: 517 KGRSRGRAKGRGRVHCHQ-NNRVNH-LGTPSSHNSILTDQVVAQPP 386
K ++GR G+GR HQ N + H + SSH + +V +PP
Sbjct: 346 KNGNKGRVVGQGRRQNHQGGNGIGHGTASSSSHPNYHPVEVSKRPP 391
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 624,875,670
Number of Sequences: 1393205
Number of extensions: 14286996
Number of successful extensions: 48724
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 40554
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48041
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31684559424
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)