Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
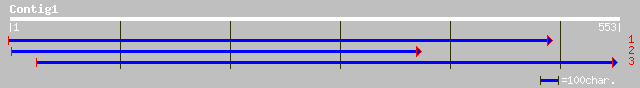
Query= KMC004729A_C01 KMC004729A_c01
(553 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
prf||2206327A T complex protein 227 8e-59
gb|AAD11431.1| T-complex protein 1 epsilon subunit [Mesembryanth... 219 2e-56
ref|NP_173859.1| T-complex chaperonin protein , epsilon subunit;... 214 6e-55
sp|P40412|TCE1_AVESA T-COMPLEX PROTEIN 1, EPSILON SUBUNIT (TCP-1... 210 8e-54
sp|P54411|TCE2_AVESA T-COMPLEX PROTEIN 1, EPSILON SUBUNIT (TCP-1... 210 8e-54
>prf||2206327A T complex protein
Length = 534
Score = 227 bits (578), Expect = 8e-59
Identities = 114/124 (91%), Positives = 120/124 (95%)
Frame = -1
Query: 553 NSIVYGGGSAEISCSVAVEAAADRFPGVEQYAIRAFADALEYVPMALAENSGLQPIETLS 374
NSIVYGGGSAEISCSVAVE AAD++PGVEQYAIRAFADAL+ VPMALAENSGLQPIETLS
Sbjct: 411 NSIVYGGGSAEISCSVAVETAADKYPGVEQYAIRAFADALDAVPMALAENSGLQPIETLS 470
Query: 373 AVKSQQIKDNNPHYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS 194
AVKSQQIK+NNP+ GIDCND+GTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS
Sbjct: 471 AVKSQQIKENNPYCGIDCNDIGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS 530
Query: 193 PFDY 182
P DY
Sbjct: 531 PSDY 534
>gb|AAD11431.1| T-complex protein 1 epsilon subunit [Mesembryanthemum crystallinum]
Length = 418
Score = 219 bits (558), Expect = 2e-56
Identities = 110/124 (88%), Positives = 119/124 (95%)
Frame = -1
Query: 553 NSIVYGGGSAEISCSVAVEAAADRFPGVEQYAIRAFADALEYVPMALAENSGLQPIETLS 374
NSIVYGGGSAEI+CS+AVEAAADR+PGVEQYAIRAFADAL+ VPMALAENSGL PIETLS
Sbjct: 295 NSIVYGGGSAEIACSLAVEAAADRYPGVEQYAIRAFADALDSVPMALAENSGLTPIETLS 354
Query: 373 AVKSQQIKDNNPHYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS 194
AVKSQQ+K+NNP GIDCNDVGTNDMREQNVFETLIGK+QQ+LLATQVVKMILKIDDVIS
Sbjct: 355 AVKSQQVKENNPCCGIDCNDVGTNDMREQNVFETLIGKKQQLLLATQVVKMILKIDDVIS 414
Query: 193 PFDY 182
P +Y
Sbjct: 415 PSEY 418
>ref|NP_173859.1| T-complex chaperonin protein , epsilon subunit; protein id:
At1g24510.1, supported by cDNA: gi_19715604 [Arabidopsis
thaliana] gi|3024697|sp|O04450|TCPE_ARATH T-complex
protein 1, epsilon subunit (TCP-1-epsilon) (CCT-epsilon)
gi|9743353|gb|AAF97977.1|AC000103_27 F21J9.17
[Arabidopsis thaliana] gi|19715605|gb|AAL91625.1|
At1g24510/F21J9_150 [Arabidopsis thaliana]
gi|23463047|gb|AAN33193.1| At1g24510/F21J9_150
[Arabidopsis thaliana]
Length = 535
Score = 214 bits (545), Expect = 6e-55
Identities = 108/123 (87%), Positives = 118/123 (95%)
Frame = -1
Query: 550 SIVYGGGSAEISCSVAVEAAADRFPGVEQYAIRAFADALEYVPMALAENSGLQPIETLSA 371
SIVYGGG+AEI+CS+AV+AAAD++PGVEQYAIRAFA+AL+ VPMALAENSGLQPIETLSA
Sbjct: 413 SIVYGGGAAEIACSLAVDAAADKYPGVEQYAIRAFAEALDSVPMALAENSGLQPIETLSA 472
Query: 370 VKSQQIKDNNPHYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVISP 191
VKSQQIK+N P YGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS
Sbjct: 473 VKSQQIKENIPFYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVISN 532
Query: 190 FDY 182
+Y
Sbjct: 533 SEY 535
>sp|P40412|TCE1_AVESA T-COMPLEX PROTEIN 1, EPSILON SUBUNIT (TCP-1-EPSILON) (CCT-EPSILON)
(TCP-K19) gi|542112|pir||S40461 t-complex-type molecular
chaperone tcp1 (clone ASTCP-K19) - oat
gi|435173|emb|CAA53396.1| T complex polypeptide 1 [Avena
sativa]
Length = 535
Score = 210 bits (535), Expect = 8e-54
Identities = 104/124 (83%), Positives = 115/124 (91%)
Frame = -1
Query: 553 NSIVYGGGSAEISCSVAVEAAADRFPGVEQYAIRAFADALEYVPMALAENSGLQPIETLS 374
NSIVYGGGSAEISCS+AVEAAADR PGVEQYAIRAFADAL+ +P+ALAENSGL PI+TL+
Sbjct: 412 NSIVYGGGSAEISCSIAVEAAADRHPGVEQYAIRAFADALDAIPLALAENSGLPPIDTLT 471
Query: 373 AVKSQQIKDNNPHYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS 194
VKSQ +K+NN GIDCNDVGTNDM+EQNVFETLIGKQQQILLATQVVKMILKIDDVI+
Sbjct: 472 VVKSQHVKENNSRCGIDCNDVGTNDMKEQNVFETLIGKQQQILLATQVVKMILKIDDVIT 531
Query: 193 PFDY 182
P +Y
Sbjct: 532 PSEY 535
>sp|P54411|TCE2_AVESA T-COMPLEX PROTEIN 1, EPSILON SUBUNIT (TCP-1-EPSILON) (CCT-EPSILON)
(TCP-K36) gi|542113|pir||S40462 t-complex-type molecular
chaperone tcp1 (clone ASTCP-K36) - oat
gi|435175|emb|CAA53397.1| t complex polypeptide 1 [Avena
sativa]
Length = 535
Score = 210 bits (535), Expect = 8e-54
Identities = 104/124 (83%), Positives = 115/124 (91%)
Frame = -1
Query: 553 NSIVYGGGSAEISCSVAVEAAADRFPGVEQYAIRAFADALEYVPMALAENSGLQPIETLS 374
NSIVYGGGSAEISCS+AVEAAADR PGVEQYAIRAFADAL+ +P+ALAENSGL PI+TL+
Sbjct: 412 NSIVYGGGSAEISCSIAVEAAADRHPGVEQYAIRAFADALDAIPLALAENSGLPPIDTLT 471
Query: 373 AVKSQQIKDNNPHYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS 194
VKSQ +K+NN GIDCNDVGTNDM+EQNVFETLIGKQQQILLATQVVKMILKIDDVI+
Sbjct: 472 VVKSQHVKENNSRCGIDCNDVGTNDMKEQNVFETLIGKQQQILLATQVVKMILKIDDVIT 531
Query: 193 PFDY 182
P +Y
Sbjct: 532 PSEY 535
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 432,221,993
Number of Sequences: 1393205
Number of extensions: 8452575
Number of successful extensions: 24656
Number of sequences better than 10.0: 379
Number of HSP's better than 10.0 without gapping: 23373
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24410
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)