Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
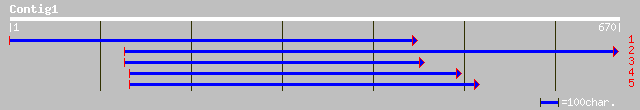
Query= KMC004721A_C01 KMC004721A_c01
(650 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196932.1| putative protein; protein id: At5g14280.1 [Arab... 184 7e-46
dbj|BAC42209.1| unknown protein [Arabidopsis thaliana] 184 7e-46
ref|NP_189224.1| hypothetical protein; protein id: At3g25950.1 [... 158 7e-38
ref|NP_189363.1| unknown protein; protein id: At3g27270.1 [Arabi... 157 9e-38
dbj|BAB01058.1| gene_id:MPE11.12~unknown protein [Arabidopsis th... 145 6e-34
>ref|NP_196932.1| putative protein; protein id: At5g14280.1 [Arabidopsis thaliana]
gi|11357556|pir||T48601 hypothetical protein F18O22.70 -
Arabidopsis thaliana gi|7573453|emb|CAB87767.1| putative
protein [Arabidopsis thaliana]
Length = 572
Score = 184 bits (468), Expect = 7e-46
Identities = 85/146 (58%), Positives = 113/146 (77%), Gaps = 4/146 (2%)
Frame = -3
Query: 648 VAHHLATLFVVVTCRHAVSHGAFAVLMLLVLAEVTSLCQNIWTLAGARRR--EDPFAAKV 475
+ HH+ATLFV +TCR V HGA A+L LL+LAEVTS CQN WTLAGAR+ E A KV
Sbjct: 427 IGHHVATLFVFLTCRFLVFHGACAILGLLILAEVTSACQNAWTLAGARKNDPESRLAVKV 486
Query: 474 YDAISPPFYAVYSVVRGLVGPYFVYRMVAFYAGGGARGLVPAWVWVSWVVVVVMAIGGSI 295
YD +SPPFYA YS+VRG++GP F +MVAFYA GGA G++P W+W+SW +VV +AI SI
Sbjct: 487 YDLLSPPFYAFYSIVRGVLGPLFFGKMVAFYARGGAHGVIPNWLWISWAIVVGIAITVSI 546
Query: 294 MWIYSLWLQLYRERSG--VKLEKKVR 223
+WI++LW++L+ ER ++++KK+R
Sbjct: 547 LWIWNLWIELFSERKANKIRVDKKIR 572
>dbj|BAC42209.1| unknown protein [Arabidopsis thaliana]
Length = 254
Score = 184 bits (468), Expect = 7e-46
Identities = 85/146 (58%), Positives = 113/146 (77%), Gaps = 4/146 (2%)
Frame = -3
Query: 648 VAHHLATLFVVVTCRHAVSHGAFAVLMLLVLAEVTSLCQNIWTLAGARRR--EDPFAAKV 475
+ HH+ATLFV +TCR V HGA A+L LL+LAEVTS CQN WTLAGAR+ E A KV
Sbjct: 109 IGHHVATLFVFLTCRFLVFHGACAILGLLILAEVTSACQNAWTLAGARKNDPESRLAVKV 168
Query: 474 YDAISPPFYAVYSVVRGLVGPYFVYRMVAFYAGGGARGLVPAWVWVSWVVVVVMAIGGSI 295
YD +SPPFYA YS+VRG++GP F +MVAFYA GGA G++P W+W+SW +VV +AI SI
Sbjct: 169 YDLLSPPFYAFYSIVRGVLGPLFFGKMVAFYARGGAHGVIPNWLWISWAIVVGIAITVSI 228
Query: 294 MWIYSLWLQLYRERSG--VKLEKKVR 223
+WI++LW++L+ ER ++++KK+R
Sbjct: 229 LWIWNLWIELFSERKANKIRVDKKIR 254
>ref|NP_189224.1| hypothetical protein; protein id: At3g25950.1 [Arabidopsis
thaliana]
Length = 251
Score = 158 bits (399), Expect = 7e-38
Identities = 70/139 (50%), Positives = 93/139 (66%)
Frame = -3
Query: 648 VAHHLATLFVVVTCRHAVSHGAFAVLMLLVLAEVTSLCQNIWTLAGARRREDPFAAKVYD 469
+ HH+ATL+V TCR AV HGA +L+LL+LAE TS CQN+WT+ G R+ + A +V +
Sbjct: 113 ILHHIATLYVFATCRFAVGHGAHGLLLLLILAEATSACQNVWTITGYRKNDVALARRVRE 172
Query: 468 AISPPFYAVYSVVRGLVGPYFVYRMVAFYAGGGARGLVPAWVWVSWVVVVVMAIGGSIMW 289
+SPPFY Y+VVRGL GP +Y M FY G A G+VP W W+SW+VV+ AI SI+W
Sbjct: 173 LLSPPFYLFYTVVRGLAGPVVLYDMATFYGSGAADGVVPRWAWLSWLVVIGFAILVSILW 232
Query: 288 IYSLWLQLYRERSGVKLEK 232
+ WL +RE + K K
Sbjct: 233 VLRNWLDWFRENNSSKKYK 251
>ref|NP_189363.1| unknown protein; protein id: At3g27270.1 [Arabidopsis thaliana]
gi|9294220|dbj|BAB02122.1| gene_id:K17E12.9~unknown
protein [Arabidopsis thaliana]
Length = 249
Score = 157 bits (398), Expect = 9e-38
Identities = 74/141 (52%), Positives = 99/141 (69%)
Frame = -3
Query: 645 AHHLATLFVVVTCRHAVSHGAFAVLMLLVLAEVTSLCQNIWTLAGARRREDPFAAKVYDA 466
AHHLA LFV +TCR+ V+HGA A+L LLV+AE TS CQN WTLA AR ++ P A ++
Sbjct: 110 AHHLAVLFVFLTCRYMVAHGACALLALLVVAEATSACQNTWTLADARGKDAPLAVSLHRF 169
Query: 465 ISPPFYAVYSVVRGLVGPYFVYRMVAFYAGGGARGLVPAWVWVSWVVVVVMAIGGSIMWI 286
++ PFYA YSV R ++ P + +M FY GGA ++P WVWVSW VV+V A+ SI+WI
Sbjct: 170 VTVPFYASYSVCRCVLAPLLIVKMTWFYVSGGADDVIPRWVWVSWTVVIVTAVTVSILWI 229
Query: 285 YSLWLQLYRERSGVKLEKKVR 223
++LW+ ++ER K KKVR
Sbjct: 230 WNLWVLFFQERYS-KFTKKVR 249
>dbj|BAB01058.1| gene_id:MPE11.12~unknown protein [Arabidopsis thaliana]
Length = 258
Score = 145 bits (365), Expect = 6e-34
Identities = 68/143 (47%), Positives = 91/143 (63%), Gaps = 1/143 (0%)
Frame = -3
Query: 648 VAHHLATLFVVVTCRHAVSHGAFAVLMLLVLAEVTSLCQNIWTLAGARRREDPFAAKVYD 469
+ HH+ATL+V TCR AV HGA +L+LL+LAE TS CQN+WT+ G R+ + A +V +
Sbjct: 113 ILHHIATLYVFATCRFAVGHGAHGLLLLLILAEATSACQNVWTITGYRKNDVALARRVRE 172
Query: 468 AISPPFYAVYSVVRGLVGPYFVYRMVAFYAGGGARGLVPAWVWVSWVVVVVMAIGGSIMW 289
+SPPFY Y+VVRGL GP +Y M FY G A G+VP W W+SW+VV+ AI
Sbjct: 173 LLSPPFYLFYTVVRGLAGPVVLYDMATFYGSGAADGVVPRWAWLSWLVVIGFAI------ 226
Query: 288 IYSLWLQLYRERSGVKL-EKKVR 223
+ S W +Y R + EK +R
Sbjct: 227 LVSCWFHVYFSRMDSPICEKLIR 249
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 652,064,382
Number of Sequences: 1393205
Number of extensions: 17136386
Number of successful extensions: 125575
Number of sequences better than 10.0: 1163
Number of HSP's better than 10.0 without gapping: 79675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 111079
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)