Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004716A_C01 KMC004716A_c01
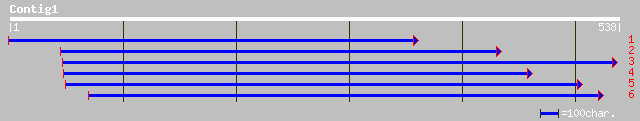
(538 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q41266|AOX2_SOYBN Alternative oxidase 2, mitochondrial precur... 235 2e-61
emb|CAC42836.1| putative alternative oxidase [Vigna unguiculata] 235 2e-61
sp|Q40294|AOX1_MANIN Alternative oxidase, mitochondrial precursor 224 4e-58
pir||S45035 alternative respiratory pathway oxidase (EC 1.-.-.-)... 224 4e-58
dbj|BAA28772.1| alternative oxidase [Oryza sativa (japonica cult... 214 7e-55
>sp|Q41266|AOX2_SOYBN Alternative oxidase 2, mitochondrial precursor
gi|7446496|pir||T08850 alternative respiratory pathway
oxidase (EC 1.-.-.-) Aox2 - soybean
gi|1946336|gb|AAB97285.1| alternative oxidase [Glycine
max]
Length = 333
Score = 235 bits (600), Expect = 2e-61
Identities = 111/119 (93%), Positives = 115/119 (96%)
Frame = -3
Query: 536 KWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENV 357
KWYER LVL VQGVFFNAFFVLY+LSPKVAHR+VGYLEEEAIHSYTEYLKD+ESGAIENV
Sbjct: 215 KWYERLLVLAVQGVFFNAFFVLYILSPKVAHRIVGYLEEEAIHSYTEYLKDLESGAIENV 274
Query: 356 PAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 180
PAPAIAIDYWRLPKDA LKDVITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 275 PAPAIAIDYWRLPKDARLKDVITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 333
>emb|CAC42836.1| putative alternative oxidase [Vigna unguiculata]
Length = 329
Score = 235 bits (600), Expect = 2e-61
Identities = 111/119 (93%), Positives = 115/119 (96%)
Frame = -3
Query: 536 KWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENV 357
KWYER LV+ VQGVFFNAFFVLY+LSPKVAHR+VGYLEEEAIHSYTEYLKDIESGAIENV
Sbjct: 211 KWYERLLVIAVQGVFFNAFFVLYILSPKVAHRIVGYLEEEAIHSYTEYLKDIESGAIENV 270
Query: 356 PAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 180
PAPAIAIDYWRLPKDA LKDVITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 271 PAPAIAIDYWRLPKDAKLKDVITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 329
>sp|Q40294|AOX1_MANIN Alternative oxidase, mitochondrial precursor
Length = 318
Score = 224 bits (572), Expect = 4e-58
Identities = 103/119 (86%), Positives = 114/119 (95%)
Frame = -3
Query: 536 KWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENV 357
KWYER LVL VQGVFFN+FFVLY+LSPK+AHR+VGYLEEEAIHSYTEYLKDI+SGAI+N+
Sbjct: 200 KWYERLLVLAVQGVFFNSFFVLYVLSPKLAHRIVGYLEEEAIHSYTEYLKDIDSGAIKNI 259
Query: 356 PAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 180
PAPAIAIDYWRLPKDATLKDVITV+RADEAHHRDVNHFASD+ GKELR+APAP+GYH
Sbjct: 260 PAPAIAIDYWRLPKDATLKDVITVVRADEAHHRDVNHFASDVQVQGKELRDAPAPVGYH 318
>pir||S45035 alternative respiratory pathway oxidase (EC 1.-.-.-) AOMI - mango
gi|488826|emb|CAA55892.1| alternative oxidase [Mangifera
indica]
Length = 274
Score = 224 bits (572), Expect = 4e-58
Identities = 103/119 (86%), Positives = 114/119 (95%)
Frame = -3
Query: 536 KWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENV 357
KWYER LVL VQGVFFN+FFVLY+LSPK+AHR+VGYLEEEAIHSYTEYLKDI+SGAI+N+
Sbjct: 156 KWYERLLVLAVQGVFFNSFFVLYVLSPKLAHRIVGYLEEEAIHSYTEYLKDIDSGAIKNI 215
Query: 356 PAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 180
PAPAIAIDYWRLPKDATLKDVITV+RADEAHHRDVNHFASD+ GKELR+APAP+GYH
Sbjct: 216 PAPAIAIDYWRLPKDATLKDVITVVRADEAHHRDVNHFASDVQVQGKELRDAPAPVGYH 274
>dbj|BAA28772.1| alternative oxidase [Oryza sativa (japonica cultivar-group)]
gi|3218546|dbj|BAA28773.1| alternative oxidase [Oryza
sativa (japonica cultivar-group)]
gi|6467126|dbj|BAA86963.1| alternative oxidase [Oryza
sativa]
Length = 332
Score = 214 bits (544), Expect = 7e-55
Identities = 97/119 (81%), Positives = 111/119 (92%)
Frame = -3
Query: 536 KWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENV 357
KWYER LV+TVQGVFFNA+F+ YLLSPK AHRVVGYLEEEAIHSYTE+LKD+E+G I+NV
Sbjct: 214 KWYERALVITVQGVFFNAYFLGYLLSPKFAHRVVGYLEEEAIHSYTEFLKDLEAGKIDNV 273
Query: 356 PAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 180
PAPAIAIDYWRLP +ATLKDV+TV+RADEAHHRDVNHFASDIH+ G EL++ PAP+GYH
Sbjct: 274 PAPAIAIDYWRLPANATLKDVVTVVRADEAHHRDVNHFASDIHYQGMELKQTPAPIGYH 332
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 458,431,629
Number of Sequences: 1393205
Number of extensions: 10087439
Number of successful extensions: 24069
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 23085
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23963
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18173652336
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)