Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004686A_C01 KMC004686A_c01
(794 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
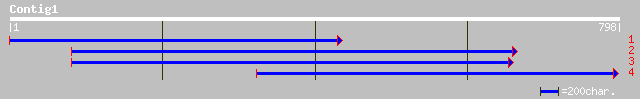
ref|NP_564053.1| Expressed protein; protein id: At1g18480.1, sup... 140 3e-32
gb|AAM60987.1| unknown [Arabidopsis thaliana] 140 3e-32
pir||E86318 protein F15H18.4 [imported] - Arabidopsis thaliana g... 110 2e-23
gb|AAL84981.1| At1g07010/F10K1_19 [Arabidopsis thaliana] 47 3e-04
ref|NP_172182.2| unknown protein; protein id: At1g07010.1, suppo... 47 3e-04
>ref|NP_564053.1| Expressed protein; protein id: At1g18480.1, supported by cDNA:
107675., supported by cDNA: gi_16226289 [Arabidopsis
thaliana] gi|16226290|gb|AAL16125.1|AF428293_1
At1g18480/F15H18_1 [Arabidopsis thaliana]
gi|23308175|gb|AAN18057.1| At1g18480/F15H18_1
[Arabidopsis thaliana]
Length = 391
Score = 140 bits (352), Expect = 3e-32
Identities = 71/112 (63%), Positives = 85/112 (75%), Gaps = 7/112 (6%)
Frame = -3
Query: 792 EAAQDCDCSTLEYVLSTVPGVKRMVMGHTIQTVGINGVCDNRAIRIDVGMSKGCGGGLPE 613
E A CDC+ LE+ LST+PGVKRM+MGHTIQ GINGVC+++AIRIDVGMSKGC GLPE
Sbjct: 283 EMAHKCDCAALEHALSTIPGVKRMIMGHTIQDAGINGVCNDKAIRIDVGMSKGCADGLPE 342
Query: 612 VLEINGNSGLRILTSNPLYQN-------AKKEKEIGLLVPEIDIPKQVEVKA 478
VLEI +SG+RI+TSNPLY+ + +GLLVP +PKQVEVKA
Sbjct: 343 VLEIRRDSGVRIVTSNPLYKENLYSHVAPDSKTGLGLLVP---VPKQVEVKA 391
>gb|AAM60987.1| unknown [Arabidopsis thaliana]
Length = 391
Score = 140 bits (352), Expect = 3e-32
Identities = 71/112 (63%), Positives = 85/112 (75%), Gaps = 7/112 (6%)
Frame = -3
Query: 792 EAAQDCDCSTLEYVLSTVPGVKRMVMGHTIQTVGINGVCDNRAIRIDVGMSKGCGGGLPE 613
E A CDC+ LE+ LST+PGVKRM+MGHTIQ GINGVC+++AIRIDVGMSKGC GLPE
Sbjct: 283 EMAHKCDCAALEHALSTIPGVKRMIMGHTIQDAGINGVCNDKAIRIDVGMSKGCADGLPE 342
Query: 612 VLEINGNSGLRILTSNPLYQN-------AKKEKEIGLLVPEIDIPKQVEVKA 478
VLEI +SG+RI+TSNPLY+ + +GLLVP +PKQVEVKA
Sbjct: 343 VLEIRRDSGVRIVTSNPLYKENPYSHVAPDSKTGLGLLVP---VPKQVEVKA 391
>pir||E86318 protein F15H18.4 [imported] - Arabidopsis thaliana
gi|6714305|gb|AAF26001.1|AC013354_20 F15H18.4
[Arabidopsis thaliana]
Length = 1702
Score = 110 bits (276), Expect = 2e-23
Identities = 52/78 (66%), Positives = 62/78 (78%)
Frame = -3
Query: 792 EAAQDCDCSTLEYVLSTVPGVKRMVMGHTIQTVGINGVCDNRAIRIDVGMSKGCGGGLPE 613
E A CDC+ LE+ LST+PGVKRM+MGHTIQ GINGVC+++AIRIDVGMSKGC GLPE
Sbjct: 283 EMAHKCDCAALEHALSTIPGVKRMIMGHTIQDAGINGVCNDKAIRIDVGMSKGCADGLPE 342
Query: 612 VLEINGNSGLRILTSNPL 559
VLEI +SG L ++ L
Sbjct: 343 VLEIRRDSGEAYLMASVL 360
>gb|AAL84981.1| At1g07010/F10K1_19 [Arabidopsis thaliana]
Length = 389
Score = 47.0 bits (110), Expect = 3e-04
Identities = 27/57 (47%), Positives = 33/57 (57%)
Frame = -3
Query: 735 GVKRMVMGHTIQTVGINGVCDNRAIRIDVGMSKGCGGGLPEVLEINGNSGLRILTSN 565
G K MV+GHT Q G+N R+DVGMS G PEVLEI G+ R++ SN
Sbjct: 321 GAKAMVVGHTPQLSGVNCEYGCGIWRVDVGMSSGVLDSRPEVLEIRGDKA-RVIRSN 376
>ref|NP_172182.2| unknown protein; protein id: At1g07010.1, supported by cDNA:
gi_19310494 [Arabidopsis thaliana]
gi|22531180|gb|AAM97094.1| unknown protein [Arabidopsis
thaliana]
Length = 389
Score = 47.0 bits (110), Expect = 3e-04
Identities = 27/57 (47%), Positives = 33/57 (57%)
Frame = -3
Query: 735 GVKRMVMGHTIQTVGINGVCDNRAIRIDVGMSKGCGGGLPEVLEINGNSGLRILTSN 565
G K MV+GHT Q G+N R+DVGMS G PEVLEI G+ R++ SN
Sbjct: 321 GAKAMVVGHTPQLSGVNCEYGCGIWRVDVGMSSGVLDSRPEVLEIRGDKA-RVIRSN 376
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 679,007,014
Number of Sequences: 1393205
Number of extensions: 15636479
Number of successful extensions: 44140
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 40980
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43841
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40055936206
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)