Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004682A_C01 KMC004682A_c01
(521 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
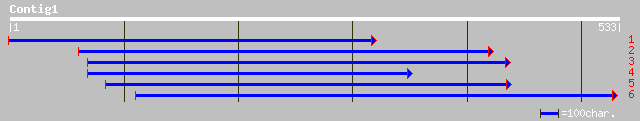
emb|CAC12824.1| putative DNAJ protein [Nicotiana tabacum] 144 5e-34
gb|AAD51625.1|AF169022_1 seed maturation protein PM37 [Glycine max] 143 1e-33
gb|AAG24642.1|AF308737_1 J1P [Daucus carota] gi|10945671|gb|AAG2... 139 2e-32
dbj|BAC53943.1| DnaJ homolog [Nicotiana tabacum] 135 2e-31
gb|AAN87055.1| tuber-induction protein [Solanum tuberosum] 134 5e-31
>emb|CAC12824.1| putative DNAJ protein [Nicotiana tabacum]
Length = 418
Score = 144 bits (364), Expect = 5e-34
Identities = 67/81 (82%), Positives = 74/81 (90%)
Frame = -3
Query: 519 FTVEFPESLSAEQVKALEAVLPPKPSSQLTDMELDECEETTLHDVNIEEETRRRQQAQQE 340
F VEFP+SL+ EQVKALEA+LPP+P SQ TDMELDECEET+LHDVNIEEE RR+Q AQQE
Sbjct: 338 FVVEFPDSLNTEQVKALEAILPPRPQSQYTDMELDECEETSLHDVNIEEEMRRKQAAQQE 397
Query: 339 AYDEDDDMHGGGAQRVQCAQQ 277
AYDEDD+MHGGG QRVQCAQQ
Sbjct: 398 AYDEDDEMHGGGGQRVQCAQQ 418
>gb|AAD51625.1|AF169022_1 seed maturation protein PM37 [Glycine max]
Length = 417
Score = 143 bits (360), Expect = 1e-33
Identities = 71/81 (87%), Positives = 76/81 (93%)
Frame = -3
Query: 519 FTVEFPESLSAEQVKALEAVLPPKPSSQLTDMELDECEETTLHDVNIEEETRRRQQAQQE 340
FTVEFP+SL+ +QVKALEAVLPPKPSSQLTDMELDECEETTLHDVN+EEETRR+QQ QE
Sbjct: 338 FTVEFPDSLNPDQVKALEAVLPPKPSSQLTDMELDECEETTLHDVNMEEETRRKQQQAQE 397
Query: 339 AYDEDDDMHGGGAQRVQCAQQ 277
AYDEDDDM GGAQRVQCAQQ
Sbjct: 398 AYDEDDDM-PGGAQRVQCAQQ 417
>gb|AAG24642.1|AF308737_1 J1P [Daucus carota] gi|10945671|gb|AAG24643.1|AF308738_1 J2P
[Daucus carota]
Length = 418
Score = 139 bits (351), Expect = 2e-32
Identities = 68/81 (83%), Positives = 75/81 (91%)
Frame = -3
Query: 519 FTVEFPESLSAEQVKALEAVLPPKPSSQLTDMELDECEETTLHDVNIEEETRRRQQAQQE 340
F+V+FPESL+ EQ KALEAVLPP+PS Q+TDMELDECEETTLHDVNIEEE RR+QQA QE
Sbjct: 339 FSVDFPESLTPEQCKALEAVLPPRPSIQMTDMELDECEETTLHDVNIEEEMRRKQQAAQE 398
Query: 339 AYDEDDDMHGGGAQRVQCAQQ 277
AYDED+DMH GGAQRVQCAQQ
Sbjct: 399 AYDEDEDMH-GGAQRVQCAQQ 418
>dbj|BAC53943.1| DnaJ homolog [Nicotiana tabacum]
Length = 339
Score = 135 bits (341), Expect = 2e-31
Identities = 66/81 (81%), Positives = 73/81 (89%)
Frame = -3
Query: 519 FTVEFPESLSAEQVKALEAVLPPKPSSQLTDMELDECEETTLHDVNIEEETRRRQQAQQE 340
FTV+FPE+LS EQ K LEAVLPPKP +Q+TDMELDECEETTLHDVNIEEE RR+QQ QE
Sbjct: 260 FTVDFPETLSLEQCKNLEAVLPPKPKTQMTDMELDECEETTLHDVNIEEEMRRKQQQAQE 319
Query: 339 AYDEDDDMHGGGAQRVQCAQQ 277
AY+ED+DMH GGAQRVQCAQQ
Sbjct: 320 AYNEDEDMH-GGAQRVQCAQQ 339
>gb|AAN87055.1| tuber-induction protein [Solanum tuberosum]
Length = 315
Score = 134 bits (338), Expect = 5e-31
Identities = 65/81 (80%), Positives = 71/81 (87%)
Frame = -3
Query: 519 FTVEFPESLSAEQVKALEAVLPPKPSSQLTDMELDECEETTLHDVNIEEETRRRQQAQQE 340
FTVEFP++LS EQ K LEAVLPPKP +Q+TDMELDECEETTLHDVNIEEE RR+QQ QE
Sbjct: 235 FTVEFPDTLSPEQCKNLEAVLPPKPKTQMTDMELDECEETTLHDVNIEEEMRRKQQQAQE 294
Query: 339 AYDEDDDMHGGGAQRVQCAQQ 277
AYDEDD+ GGAQRVQCAQQ
Sbjct: 295 AYDEDDEDMHGGAQRVQCAQQ 315
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 450,060,163
Number of Sequences: 1393205
Number of extensions: 9546440
Number of successful extensions: 35701
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 28380
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33101
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)