Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004673A_C01 KMC004673A_c01
(577 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
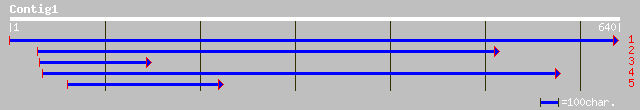
ref|NP_566300.1| expressed protein; protein id: At3g07170.1, sup... 113 1e-24
ref|NP_199679.1| putative protein; protein id: At5g48680.1 [Arab... 88 9e-17
ref|NP_177175.2| hypothetical protein; protein id: At1g70180.1, ... 62 7e-09
ref|NP_009121.1| Sec23-interacting protein p125 [Homo sapiens] g... 49 3e-05
gb|AAO15299.1| MSTP053 [Homo sapiens] 49 5e-05
>ref|NP_566300.1| expressed protein; protein id: At3g07170.1, supported by cDNA:
gi_15294217 [Arabidopsis thaliana]
gi|6642648|gb|AAF20229.1|AC012395_16 unknown protein
[Arabidopsis thaliana]
gi|15294218|gb|AAK95286.1|AF410300_1 AT3g07170/T1B9_17
[Arabidopsis thaliana] gi|22655432|gb|AAM98308.1|
At3g07170/T1B9_17 [Arabidopsis thaliana]
Length = 203
Score = 113 bits (283), Expect = 1e-24
Identities = 64/108 (59%), Positives = 80/108 (73%), Gaps = 1/108 (0%)
Frame = -2
Query: 576 QPKNSDPPKSKV-MGKPSNKSVGVEAPAAKIKRSTNPAPKTLARKAETSVDEFLQSLGLE 400
QPKNSDPPKSK +PS KSV E + +++++ A + +++A++SVD FL+SLGLE
Sbjct: 102 QPKNSDPPKSKAEAARPSMKSVATET---ETRKTSSQATRKKSQQADSSVDSFLESLGLE 158
Query: 399 KYLISFQAEELLFQVDMTALNHMTDEDLKAMGIPMGPRKKILLALESK 256
KY +FQ EE VDM AL HMTD+DLKAM IPMGPRKKILLAL SK
Sbjct: 159 KYSTAFQVEE----VDMDALMHMTDDDLKAMLIPMGPRKKILLALGSK 202
>ref|NP_199679.1| putative protein; protein id: At5g48680.1 [Arabidopsis thaliana]
Length = 183
Score = 87.8 bits (216), Expect = 9e-17
Identities = 52/93 (55%), Positives = 60/93 (63%), Gaps = 8/93 (8%)
Frame = -2
Query: 519 SVGVEAPAAKIKRSTNPAPKTLARKA--------ETSVDEFLQSLGLEKYLISFQAEELL 364
SV V K+ R+ NP K R+A + SVD FL+SLGLEKY +FQ EE
Sbjct: 90 SVSVADLRDKLSRTVNPQTKNSKREAVRPAIKKNDASVDSFLESLGLEKYSTAFQVEE-- 147
Query: 363 FQVDMTALNHMTDEDLKAMGIPMGPRKKILLAL 265
VDM AL HMTD+DLKA+ IPMGPRKKILLAL
Sbjct: 148 --VDMDALRHMTDDDLKALLIPMGPRKKILLAL 178
>ref|NP_177175.2| hypothetical protein; protein id: At1g70180.1, supported by cDNA:
gi_17473773 [Arabidopsis thaliana]
gi|17473774|gb|AAL38323.1| unknown protein [Arabidopsis
thaliana]
Length = 460
Score = 61.6 bits (148), Expect = 7e-09
Identities = 33/59 (55%), Positives = 41/59 (68%)
Frame = -2
Query: 435 SVDEFLQSLGLEKYLISFQAEELLFQVDMTALNHMTDEDLKAMGIPMGPRKKILLALES 259
SVD FL S+GL KY ++F+ EE VDMT + M + DLK + IPMGPRKKIL A+ S
Sbjct: 402 SVDGFLNSIGLGKYSLAFKREE----VDMTTIKQMKESDLKDLIIPMGPRKKILQAIAS 456
>ref|NP_009121.1| Sec23-interacting protein p125 [Homo sapiens]
gi|4760647|dbj|BAA77392.1| phospholipase [Homo sapiens]
Length = 1000
Score = 49.3 bits (116), Expect = 3e-05
Identities = 27/64 (42%), Positives = 40/64 (62%)
Frame = -2
Query: 447 KAETSVDEFLQSLGLEKYLISFQAEELLFQVDMTALNHMTDEDLKAMGIPMGPRKKILLA 268
K ++ E L++L L +Y +F+ E+ +DM +L T +DLK MGIP+GPRKKI
Sbjct: 644 KEVLTLQETLEALSLSEYFSTFEKEK----IDMESLLMCTVDDLKEMGIPLGPRKKIANF 699
Query: 267 LESK 256
+E K
Sbjct: 700 VEHK 703
>gb|AAO15299.1| MSTP053 [Homo sapiens]
Length = 924
Score = 48.9 bits (115), Expect = 5e-05
Identities = 26/60 (43%), Positives = 39/60 (64%)
Frame = -2
Query: 435 SVDEFLQSLGLEKYLISFQAEELLFQVDMTALNHMTDEDLKAMGIPMGPRKKILLALESK 256
++ E L++L L +Y +F+ E+ +DM +L T +DLK MGIP+GPRKKI +E K
Sbjct: 648 TLQETLEALSLSEYFSTFEKEK----IDMESLLMCTVDDLKEMGIPLGPRKKIANFVEHK 703
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,188,088
Number of Sequences: 1393205
Number of extensions: 9923735
Number of successful extensions: 35428
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 33752
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35340
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)