Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
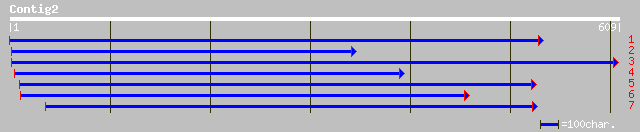
Query= KMC004641A_C02 KMC004641A_c02
(591 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA82306.1| peroxidase [Nicotiana tabacum] 168 2e-50
emb|CAD67477.1| peroxidase [Asparagus officinalis] 155 4e-44
gb|AAA96137.1| peroxidase 147 3e-42
gb|AAL93151.1|AF485265_1 gaiacol peroxidase [Gossypium hirsutum] 142 6e-42
emb|CAD67478.1| peroxidase [Asparagus officinalis] 145 1e-41
>dbj|BAA82306.1| peroxidase [Nicotiana tabacum]
Length = 321
Score = 168 bits (425), Expect(3) = 2e-50
Identities = 79/100 (79%), Positives = 87/100 (87%)
Frame = -1
Query: 591 VALSGAHTIGQARCTSFRARIYNETSTIESSFATSRKSNCPSTSGSGDNNLAPLDLQTPT 412
VALSGAHTIGQARCTSFRARIYNET+ +++SFA +R+SNCP +SGSGDNNLAPLDLQTP
Sbjct: 187 VALSGAHTIGQARCTSFRARIYNETNNLDASFARTRQSNCPRSSGSGDNNLAPLDLQTPN 246
Query: 411 SFDNNYFKNLVQNKGLLHSDQQLFNGGSTDSTVRGYRHQP 292
FDNNYFKNLV KGLLHSDQQLFNGGS DS V Y + P
Sbjct: 247 KFDNNYFKNLVDKKGLLHSDQQLFNGGSADSIVTSYSNNP 286
Score = 51.2 bits (121), Expect(3) = 2e-50
Identities = 22/26 (84%), Positives = 24/26 (91%)
Frame = -3
Query: 262 AMVKMGDISPLTGSNGEIRKNCRKTN 185
AM+KMGDI PLTGSNGEIRKNCR+ N
Sbjct: 296 AMIKMGDIRPLTGSNGEIRKNCRRLN 321
Score = 22.7 bits (47), Expect(3) = 2e-50
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = -2
Query: 296 NPSSFSSDF 270
NPSSFSSDF
Sbjct: 285 NPSSFSSDF 293
>emb|CAD67477.1| peroxidase [Asparagus officinalis]
Length = 315
Score = 155 bits (391), Expect(2) = 4e-44
Identities = 75/96 (78%), Positives = 84/96 (87%)
Frame = -1
Query: 591 VALSGAHTIGQARCTSFRARIYNETSTIESSFATSRKSNCPSTSGSGDNNLAPLDLQTPT 412
VALSGAHTIGQARCTSFR+ IYN+ S I+ SFAT RKSNCP SGSGD NLAPLDLQTPT
Sbjct: 182 VALSGAHTIGQARCTSFRSHIYND-SDIDPSFATLRKSNCPKQSGSGDMNLAPLDLQTPT 240
Query: 411 SFDNNYFKNLVQNKGLLHSDQQLFNGGSTDSTVRGY 304
+FDNNY++NLV KGL+HSDQ+LFNGGSTDS V+ Y
Sbjct: 241 TFDNNYYRNLVVKKGLMHSDQELFNGGSTDSLVKSY 276
Score = 45.1 bits (105), Expect(2) = 4e-44
Identities = 19/25 (76%), Positives = 21/25 (84%)
Frame = -3
Query: 259 MVKMGDISPLTGSNGEIRKNCRKTN 185
M+KMGD+SPL GSNGEIRK C K N
Sbjct: 291 MIKMGDVSPLVGSNGEIRKICSKVN 315
>gb|AAA96137.1| peroxidase
Length = 136
Score = 147 bits (371), Expect(2) = 3e-42
Identities = 70/96 (72%), Positives = 81/96 (83%)
Frame = -1
Query: 591 VALSGAHTIGQARCTSFRARIYNETSTIESSFATSRKSNCPSTSGSGDNNLAPLDLQTPT 412
VALSG+HTIGQARCTSFR IYN+T I+ SFA R+ NCP SG+GD+NLAPLDLQTPT
Sbjct: 3 VALSGSHTIGQARCTSFRGHIYNDTD-IDPSFAKLRQKNCPRQSGTGDSNLAPLDLQTPT 61
Query: 411 SFDNNYFKNLVQNKGLLHSDQQLFNGGSTDSTVRGY 304
F+NNY+KNL+ KGLLHSDQ+LFNGGSTDS V+ Y
Sbjct: 62 HFENNYYKNLINKKGLLHSDQELFNGGSTDSLVQTY 97
Score = 46.6 bits (109), Expect(2) = 3e-42
Identities = 21/28 (75%), Positives = 24/28 (85%)
Frame = -3
Query: 268 PGAMVKMGDISPLTGSNGEIRKNCRKTN 185
PG M+KMGD+ PLTGS GEIRKNCR+ N
Sbjct: 110 PG-MIKMGDLLPLTGSKGEIRKNCRRMN 136
>gb|AAL93151.1|AF485265_1 gaiacol peroxidase [Gossypium hirsutum]
Length = 320
Score = 142 bits (359), Expect(2) = 6e-42
Identities = 69/96 (71%), Positives = 81/96 (83%)
Frame = -1
Query: 591 VALSGAHTIGQARCTSFRARIYNETSTIESSFATSRKSNCPSTSGSGDNNLAPLDLQTPT 412
VALSGAHTIG+ARC FR RIYN+T I++SFA +R+S+CP T GSGDNNLAPLDL TP
Sbjct: 187 VALSGAHTIGKARCLVFRNRIYNDT-IIDTSFAKTRRSSCPRTRGSGDNNLAPLDLATPN 245
Query: 411 SFDNNYFKNLVQNKGLLHSDQQLFNGGSTDSTVRGY 304
SFD+ YF+NL+ KGLLHSDQ+LFNGGSTDS V+ Y
Sbjct: 246 SFDSKYFENLLNKKGLLHSDQELFNGGSTDSLVKTY 281
Score = 50.1 bits (118), Expect(2) = 6e-42
Identities = 22/26 (84%), Positives = 23/26 (87%)
Frame = -3
Query: 262 AMVKMGDISPLTGSNGEIRKNCRKTN 185
AM+KMGDI PLTGSNGEIRKNC K N
Sbjct: 295 AMIKMGDIKPLTGSNGEIRKNCGKPN 320
>emb|CAD67478.1| peroxidase [Asparagus officinalis]
Length = 301
Score = 145 bits (366), Expect(2) = 1e-41
Identities = 69/96 (71%), Positives = 80/96 (82%)
Frame = -1
Query: 591 VALSGAHTIGQARCTSFRARIYNETSTIESSFATSRKSNCPSTSGSGDNNLAPLDLQTPT 412
VALSGAHTIGQARCTSFR IYN+ I++SFA+ R+ CP SGSGD NLAPLDLQTPT
Sbjct: 168 VALSGAHTIGQARCTSFRGHIYNDAD-IDASFASLRQKICPRKSGSGDTNLAPLDLQTPT 226
Query: 411 SFDNNYFKNLVQNKGLLHSDQQLFNGGSTDSTVRGY 304
+FDNNY+KNL+ KGLLHSDQ+LFN G+TDS V+ Y
Sbjct: 227 AFDNNYYKNLINKKGLLHSDQELFNNGATDSLVKSY 262
Score = 46.2 bits (108), Expect(2) = 1e-41
Identities = 21/26 (80%), Positives = 22/26 (83%)
Frame = -3
Query: 262 AMVKMGDISPLTGSNGEIRKNCRKTN 185
AM+KMGDISPLTGS GEIRK C K N
Sbjct: 276 AMIKMGDISPLTGSKGEIRKICSKIN 301
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 545,487,894
Number of Sequences: 1393205
Number of extensions: 12626402
Number of successful extensions: 44894
Number of sequences better than 10.0: 469
Number of HSP's better than 10.0 without gapping: 38998
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43956
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)