Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004634A_C01 KMC004634A_c01
(556 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
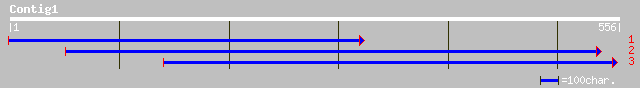
ref|NP_178317.1| putative C2H2-type zinc finger protein; protein... 47 1e-04
dbj|BAC42382.1| putative C2H2-type zinc finger protein [Arabidop... 47 1e-04
ref|NP_172910.2| zinc finger protein, putative; protein id: At1g... 38 0.095
gb|AAF63168.1|AC010657_4 T5E21.8 [Arabidopsis thaliana] 38 0.095
ref|NP_648260.1| CG13312-PA [Drosophila melanogaster] gi|7295034... 33 2.3
>ref|NP_178317.1| putative C2H2-type zinc finger protein; protein id: At2g02080.1
[Arabidopsis thaliana] gi|25410961|pir||G84432 probable
C2H2-type zinc finger protein [imported] - Arabidopsis
thaliana gi|4038045|gb|AAC97227.1| putative C2H2-type
zinc finger protein [Arabidopsis thaliana]
Length = 439
Score = 47.4 bits (111), Expect = 1e-04
Identities = 23/34 (67%), Positives = 26/34 (75%), Gaps = 6/34 (17%)
Frame = -3
Query: 401 DLSFG------GSDKLTLDFLGVGGMMRNINSGG 318
D+ FG GSDKLTLDFLGVGGM+RN+N GG
Sbjct: 376 DVQFGDNGNMSGSDKLTLDFLGVGGMVRNVNRGG 409
>dbj|BAC42382.1| putative C2H2-type zinc finger protein [Arabidopsis thaliana]
gi|29028906|gb|AAO64832.1| At2g02080 [Arabidopsis
thaliana]
Length = 516
Score = 47.4 bits (111), Expect = 1e-04
Identities = 23/34 (67%), Positives = 26/34 (75%), Gaps = 6/34 (17%)
Frame = -3
Query: 401 DLSFG------GSDKLTLDFLGVGGMMRNINSGG 318
D+ FG GSDKLTLDFLGVGGM+RN+N GG
Sbjct: 453 DVQFGDNGNMSGSDKLTLDFLGVGGMVRNVNRGG 486
>ref|NP_172910.2| zinc finger protein, putative; protein id: At1g14580.1, supported
by cDNA: gi_20259483 [Arabidopsis thaliana]
gi|20259484|gb|AAM13862.1| putative zinc finger protein
[Arabidopsis thaliana] gi|22136762|gb|AAM91700.1|
putative zinc finger protein [Arabidopsis thaliana]
Length = 467
Score = 37.7 bits (86), Expect = 0.095
Identities = 18/29 (62%), Positives = 23/29 (79%), Gaps = 1/29 (3%)
Frame = -3
Query: 401 DLSFGGSDKLTLDFLGV-GGMMRNINSGG 318
+LS GGSD+LTLDFLGV GG++ +N G
Sbjct: 415 NLSMGGSDRLTLDFLGVNGGIVSTVNGRG 443
>gb|AAF63168.1|AC010657_4 T5E21.8 [Arabidopsis thaliana]
Length = 499
Score = 37.7 bits (86), Expect = 0.095
Identities = 18/29 (62%), Positives = 23/29 (79%), Gaps = 1/29 (3%)
Frame = -3
Query: 401 DLSFGGSDKLTLDFLGV-GGMMRNINSGG 318
+LS GGSD+LTLDFLGV GG++ +N G
Sbjct: 447 NLSMGGSDRLTLDFLGVNGGIVSTVNGRG 475
>ref|NP_648260.1| CG13312-PA [Drosophila melanogaster] gi|7295034|gb|AAF50361.1|
CG13312-PA [Drosophila melanogaster]
Length = 342
Score = 33.1 bits (74), Expect = 2.3
Identities = 18/65 (27%), Positives = 32/65 (48%)
Frame = +2
Query: 362 NPTSTCQNLQNSNQDFVATFHSLACYQIMDCVVPVGNRVDHQSVKIAAVIFHVCSHYHIH 541
NP + CQ +Q++ V T S C+Q +DC + G + ++ ++ S +
Sbjct: 262 NPEAYCQAMQSAGYYPVGTDASTTCHQYIDCFLNGGTWGGNMYTCPGSLYYNSASRTCVS 321
Query: 542 TLPSS 556
TLPS+
Sbjct: 322 TLPST 326
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 463,574,297
Number of Sequences: 1393205
Number of extensions: 9692639
Number of successful extensions: 23747
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 22931
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23730
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)