Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004627A_C01 KMC004627A_c01
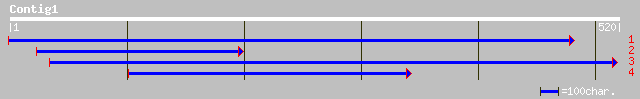
(519 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196320.1| nucleoid DNA-binding-like protein; protein id: ... 104 5e-22
ref|NP_191008.1| nucleoid DNA-binding - like protein; protein id... 104 7e-22
gb|AAM66983.1| nucleoid DNA-binding-like protein [Arabidopsis th... 104 7e-22
dbj|BAC15479.1| similar to nucleoid DNA-binding-like protein [Or... 100 1e-20
emb|CAA06698.1| hypothetical protein [Cicer arietinum] 92 5e-18
>ref|NP_196320.1| nucleoid DNA-binding-like protein; protein id: At5g07030.1,
supported by cDNA: 15560. [Arabidopsis thaliana]
gi|9759559|dbj|BAB11161.1| nucleoid DNA-binding-like
protein [Arabidopsis thaliana]
gi|21553652|gb|AAM62745.1| nucleoid DNA-binding-like
protein [Arabidopsis thaliana]
Length = 439
Score = 104 bits (260), Expect = 5e-22
Identities = 48/62 (77%), Positives = 57/62 (91%)
Frame = -2
Query: 518 TLPQDNLLIHSTAGSTTCLAMAGAPDNVNSVLNVIANMQQQNHRVLYDVPNSRLGVAREL 339
T+P DNL++HSTAGST+CLAMA AP+NVNSV+NVIA+MQQQNHRVL DVPN RLG+ARE
Sbjct: 378 TMPADNLMLHSTAGSTSCLAMAAAPENVNSVVNVIASMQQQNHRVLIDVPNGRLGLARER 437
Query: 338 CT 333
C+
Sbjct: 438 CS 439
>ref|NP_191008.1| nucleoid DNA-binding - like protein; protein id: At3g54400.1,
supported by cDNA: 8987., supported by cDNA: gi_17979256
[Arabidopsis thaliana] gi|11358589|pir||T47599 nucleoid
DNA-binding-like protein - Arabidopsis thaliana
gi|7288018|emb|CAB81805.1| nucleoid DNA-binding-like
protein [Arabidopsis thaliana]
gi|17979257|gb|AAL49945.1| AT3g54400/T12E18_90
[Arabidopsis thaliana] gi|21700851|gb|AAM70549.1|
AT3g54400/T12E18_90 [Arabidopsis thaliana]
Length = 425
Score = 104 bits (259), Expect = 7e-22
Identities = 50/62 (80%), Positives = 56/62 (89%)
Frame = -2
Query: 518 TLPQDNLLIHSTAGSTTCLAMAGAPDNVNSVLNVIANMQQQNHRVLYDVPNSRLGVAREL 339
TLP DNLLIHS+AG+ +CLAMA AP NVNSVLNVIA+MQQQNHRVL DVPNSRLG++RE
Sbjct: 364 TLPPDNLLIHSSAGNLSCLAMAAAPVNVNSVLNVIASMQQQNHRVLIDVPNSRLGISRET 423
Query: 338 CT 333
CT
Sbjct: 424 CT 425
>gb|AAM66983.1| nucleoid DNA-binding-like protein [Arabidopsis thaliana]
Length = 425
Score = 104 bits (259), Expect = 7e-22
Identities = 50/62 (80%), Positives = 56/62 (89%)
Frame = -2
Query: 518 TLPQDNLLIHSTAGSTTCLAMAGAPDNVNSVLNVIANMQQQNHRVLYDVPNSRLGVAREL 339
TLP DNLLIHS+AG+ +CLAMA AP NVNSVLNVIA+MQQQNHRVL DVPNSRLG++RE
Sbjct: 364 TLPPDNLLIHSSAGNLSCLAMAAAPVNVNSVLNVIASMQQQNHRVLIDVPNSRLGISRET 423
Query: 338 CT 333
CT
Sbjct: 424 CT 425
>dbj|BAC15479.1| similar to nucleoid DNA-binding-like protein [Oryza sativa
(japonica cultivar-group)] gi|22831290|dbj|BAC16145.1|
similar to nucleoid DNA-binding-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 449
Score = 100 bits (248), Expect = 1e-20
Identities = 45/62 (72%), Positives = 56/62 (89%)
Frame = -2
Query: 518 TLPQDNLLIHSTAGSTTCLAMAGAPDNVNSVLNVIANMQQQNHRVLYDVPNSRLGVAREL 339
TLP++N++IH+T G+T+CLAMA APD VN+VLNVIA+MQQQNHRVL+DVPN R+G ARE
Sbjct: 386 TLPEENVVIHTTYGTTSCLAMAAAPDGVNTVLNVIASMQQQNHRVLFDVPNGRVGFARES 445
Query: 338 CT 333
CT
Sbjct: 446 CT 447
>emb|CAA06698.1| hypothetical protein [Cicer arietinum]
Length = 99
Score = 91.7 bits (226), Expect = 5e-18
Identities = 43/61 (70%), Positives = 51/61 (83%)
Frame = -2
Query: 518 TLPQDNLLIHSTAGSTTCLAMAGAPDNVNSVLNVIANMQQQNHRVLYDVPNSRLGVAREL 339
TLP +N LIHS++GS CLAMA AP NVNSVLNVIAN QQQN RVL+D N+++G+AREL
Sbjct: 38 TLPMENSLIHSSSGSLACLAMAAAPSNVNSVLNVIANFQQQNLRVLFDTVNNKVGIAREL 97
Query: 338 C 336
C
Sbjct: 98 C 98
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,508,164
Number of Sequences: 1393205
Number of extensions: 8496693
Number of successful extensions: 21273
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 20491
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21257
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16442828304
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)