Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004624A_C02 KMC004624A_c02
(705 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
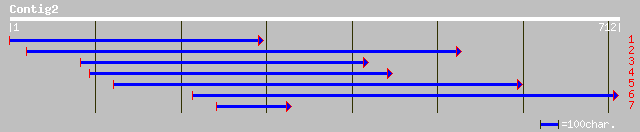
gb|AAC32134.1| KIAA0107-like protein [Picea mariana] 255 5e-67
sp|Q8W425|PSD6_ORYSA 26S proteasome non-ATPase regulatory subuni... 254 6e-67
ref|NP_567709.1| 26S proteasome regulatory subunit (RPN7), putat... 251 7e-66
pir||T06666 26S proteasome regulatory particle chain RPN7 homolo... 238 5e-62
emb|CAC09486.1| contains similarity to F6I7.30 [Oryza sativa (in... 213 2e-54
>gb|AAC32134.1| KIAA0107-like protein [Picea mariana]
Length = 232
Score = 255 bits (651), Expect = 5e-67
Identities = 124/138 (89%), Positives = 133/138 (95%)
Frame = -3
Query: 703 KIPHLSEFMNSLYDCQYKSFFSAFAGLTEQVKLDRYLHPHFRYYMREVRTVVYSQFLESY 524
KIPHLS+F+NSLY CQYKSFF AFAGLT+Q++LDRYLHPHFRYYMREVRTVVYSQFLESY
Sbjct: 95 KIPHLSDFLNSLYGCQYKSFFVAFAGLTDQIRLDRYLHPHFRYYMREVRTVVYSQFLESY 154
Query: 523 KSVTIEAMPKAFGVTVDFIDVELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIK 344
KSVT+EAM +AFGV+VDFID ELSRFIAAG+LHCKIDKV GVLETNRPDAKNALYQATIK
Sbjct: 155 KSVTMEAMARAFGVSVDFIDRELSRFIAAGRLHCKIDKVVGVLETNRPDAKNALYQATIK 214
Query: 343 QGDFLLNRIQKLSRVIDL 290
QGDFLLNRIQKLSRVIDL
Sbjct: 215 QGDFLLNRIQKLSRVIDL 232
>sp|Q8W425|PSD6_ORYSA 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome
regulatory particle non-ATPase subunit 7) (OsRPN7)
gi|17297977|dbj|BAB78486.1| 26S proteasome regulatory
particle non-ATPase subunit7 [Oryza sativa (japonica
cultivar-group)] gi|21741449|emb|CAD41393.1|
OJ000223_09.5 [Oryza sativa (japonica cultivar-group)]
Length = 389
Score = 254 bits (650), Expect = 6e-67
Identities = 124/138 (89%), Positives = 133/138 (95%)
Frame = -3
Query: 703 KIPHLSEFMNSLYDCQYKSFFSAFAGLTEQVKLDRYLHPHFRYYMREVRTVVYSQFLESY 524
K+PHLSEF+NSLY+CQYKSFF+AF+GLTEQ+KLDRYL PHFRYYMREVRTVVYSQFLESY
Sbjct: 252 KVPHLSEFLNSLYNCQYKSFFAAFSGLTEQIKLDRYLQPHFRYYMREVRTVVYSQFLESY 311
Query: 523 KSVTIEAMPKAFGVTVDFIDVELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIK 344
KSVT+EAM AFGVTVDFID+ELSRFIAAGKLHCKIDKVA VLETNRPDA+NA YQATIK
Sbjct: 312 KSVTMEAMASAFGVTVDFIDLELSRFIAAGKLHCKIDKVACVLETNRPDARNAFYQATIK 371
Query: 343 QGDFLLNRIQKLSRVIDL 290
QGDFLLNRIQKLSRVIDL
Sbjct: 372 QGDFLLNRIQKLSRVIDL 389
>ref|NP_567709.1| 26S proteasome regulatory subunit (RPN7), putative; protein id:
At4g24820.1, supported by cDNA: 38927., supported by
cDNA: gi_15450839, supported by cDNA: gi_20259841
[Arabidopsis thaliana] gi|20978551|sp|Q93Y35|PSD6_ARATH
Probable 26S proteasome non-ATPase regulatory subunit 6
gi|15450840|gb|AAK96691.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
gi|20259842|gb|AAM13268.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
gi|21593433|gb|AAM65400.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
gi|23397029|gb|AAN31800.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
Length = 387
Score = 251 bits (641), Expect = 7e-66
Identities = 123/138 (89%), Positives = 132/138 (95%)
Frame = -3
Query: 703 KIPHLSEFMNSLYDCQYKSFFSAFAGLTEQVKLDRYLHPHFRYYMREVRTVVYSQFLESY 524
KIP LSEF+NSLY+CQYK+FFSAFAG+ Q+K DRYL+PHFR+YMREVRTVVYSQFLESY
Sbjct: 250 KIPFLSEFLNSLYECQYKAFFSAFAGMAVQIKYDRYLYPHFRFYMREVRTVVYSQFLESY 309
Query: 523 KSVTIEAMPKAFGVTVDFIDVELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIK 344
KSVT+EAM KAFGV+VDFID ELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIK
Sbjct: 310 KSVTVEAMAKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIK 369
Query: 343 QGDFLLNRIQKLSRVIDL 290
QGDFLLNRIQKLSRVIDL
Sbjct: 370 QGDFLLNRIQKLSRVIDL 387
>pir||T06666 26S proteasome regulatory particle chain RPN7 homolog F6I7.30 -
Arabidopsis thaliana gi|4678261|emb|CAB41122.1| putative
proteasome regulatory subunit [Arabidopsis thaliana]
gi|7269333|emb|CAB79392.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
Length = 406
Score = 238 bits (608), Expect = 5e-62
Identities = 122/157 (77%), Positives = 132/157 (83%), Gaps = 19/157 (12%)
Frame = -3
Query: 703 KIPHLSEFMNSLYDCQYKSFFSAF-------------------AGLTEQVKLDRYLHPHF 581
KIP LSEF+NSLY+CQYK+FFSAF +G+ Q+K DRYL+PHF
Sbjct: 250 KIPFLSEFLNSLYECQYKAFFSAFGKFITRTRGHTSECMLLGFSGMAVQIKYDRYLYPHF 309
Query: 580 RYYMREVRTVVYSQFLESYKSVTIEAMPKAFGVTVDFIDVELSRFIAAGKLHCKIDKVAG 401
R+YMREVRTVVYSQFLESYKSVT+EAM KAFGV+VDFID ELSRFIAAGKLHCKIDKVAG
Sbjct: 310 RFYMREVRTVVYSQFLESYKSVTVEAMAKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAG 369
Query: 400 VLETNRPDAKNALYQATIKQGDFLLNRIQKLSRVIDL 290
VLETNRPDAKNALYQATIKQGDFLLNRIQKLSRVIDL
Sbjct: 370 VLETNRPDAKNALYQATIKQGDFLLNRIQKLSRVIDL 406
>emb|CAC09486.1| contains similarity to F6I7.30 [Oryza sativa (indica
cultivar-group)]
Length = 209
Score = 213 bits (543), Expect = 2e-54
Identities = 106/114 (92%), Positives = 110/114 (95%)
Frame = -3
Query: 631 AGLTEQVKLDRYLHPHFRYYMREVRTVVYSQFLESYKSVTIEAMPKAFGVTVDFIDVELS 452
AGLTEQ+KLDRYL PHFRYYMREVRTVVYSQFLESYKSVT+EAM AFGVTVDFID+ELS
Sbjct: 96 AGLTEQIKLDRYLQPHFRYYMREVRTVVYSQFLESYKSVTMEAMASAFGVTVDFIDLELS 155
Query: 451 RFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIKQGDFLLNRIQKLSRVIDL 290
RFIAAGKLHCKIDKVAGVLETNRPDA+NA YQATIKQGDFLLNRIQKLSRVIDL
Sbjct: 156 RFIAAGKLHCKIDKVAGVLETNRPDARNAFYQATIKQGDFLLNRIQKLSRVIDL 209
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 581,978,413
Number of Sequences: 1393205
Number of extensions: 11852991
Number of successful extensions: 24152
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 23556
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24143
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32091529758
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)