Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004604A_C01 KMC004604A_c01
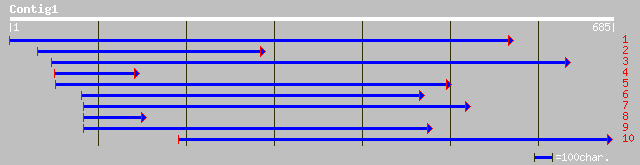
(685 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_195928.1| putative protein; protein id: At5g03080.1 [Arab... 115 6e-25
pir||T48329 hypothetical protein F15A17.110 - Arabidopsis thalia... 114 1e-24
gb|AAO51881.1| similar to Mus musculus (Mouse). RIKEN cDNA 06100... 69 5e-11
gb|EAA08176.1| ebiP2778 [Anopheles gambiae str. PEST] 68 1e-10
dbj|BAB27187.1| unnamed protein product [Mus musculus] 66 5e-10
>ref|NP_195928.1| putative protein; protein id: At5g03080.1 [Arabidopsis thaliana]
Length = 226
Score = 115 bits (288), Expect = 6e-25
Identities = 57/83 (68%), Positives = 67/83 (80%)
Frame = -3
Query: 683 VYLGYHTLAQVFAGTILGVSLGAIWFWVVNTVLRPYFSLIEESEFGKRFYVKDTSHIPNV 504
VYLGYHT+AQVFAG LG +GA WFWVVN+VL P+F +IEES G+ YVKDTSHIP+V
Sbjct: 146 VYLGYHTVAQVFAGAALGGIVGASWFWVVNSVLYPFFPVIEESVLGRWLYVKDTSHIPDV 205
Query: 503 LKFEYEQARAARKNVPSTSNKSD 435
LKFEY+ ARAARK++ S KSD
Sbjct: 206 LKFEYDNARAARKDMDSA--KSD 226
>pir||T48329 hypothetical protein F15A17.110 - Arabidopsis thaliana
gi|7413585|emb|CAB86075.1| putative protein [Arabidopsis
thaliana]
Length = 268
Score = 114 bits (286), Expect = 1e-24
Identities = 54/77 (70%), Positives = 64/77 (82%)
Frame = -3
Query: 683 VYLGYHTLAQVFAGTILGVSLGAIWFWVVNTVLRPYFSLIEESEFGKRFYVKDTSHIPNV 504
VYLGYHT+AQVFAG LG +GA WFWVVN+VL P+F +IEES G+ YVKDTSHIP+V
Sbjct: 146 VYLGYHTVAQVFAGAALGGIVGASWFWVVNSVLYPFFPVIEESVLGRWLYVKDTSHIPDV 205
Query: 503 LKFEYEQARAARKNVPS 453
LKFEY+ ARAARK++ S
Sbjct: 206 LKFEYDNARAARKDMDS 222
>gb|AAO51881.1| similar to Mus musculus (Mouse). RIKEN cDNA 0610011H20 gene
[Dictyostelium discoideum]
Length = 229
Score = 69.3 bits (168), Expect = 5e-11
Identities = 33/79 (41%), Positives = 47/79 (58%), Gaps = 1/79 (1%)
Frame = -3
Query: 683 VYLGYHTLAQVFAGTILGVSLGAIWFWVVNTVLRPY-FSLIEESEFGKRFYVKDTSHIPN 507
V+L YHT QVF G+ +G+ LG IW+ V+ + RPY F +I GK FY++D+S I +
Sbjct: 145 VHLYYHTAKQVFCGSFIGICLGFIWYGVIEYIFRPYLFPIIINHPIGKYFYLRDSSEIED 204
Query: 506 VLKFEYEQARAARKNVPST 450
+L FEY K + T
Sbjct: 205 LLNFEYTNVMNKVKTINKT 223
>gb|EAA08176.1| ebiP2778 [Anopheles gambiae str. PEST]
Length = 221
Score = 67.8 bits (164), Expect = 1e-10
Identities = 30/69 (43%), Positives = 45/69 (64%)
Frame = -3
Query: 683 VYLGYHTLAQVFAGTILGVSLGAIWFWVVNTVLRPYFSLIEESEFGKRFYVKDTSHIPNV 504
+YL YHTL+QV G ++G +GA+WF + + VL PYF ++ + F ++DT+ IPN+
Sbjct: 143 IYLLYHTLSQVLIGALVGTVMGALWFLLTHFVLTPYFPMVVLWRVSELFLLRDTTLIPNI 202
Query: 503 LKFEYEQAR 477
L FEY R
Sbjct: 203 LWFEYTVTR 211
>dbj|BAB27187.1| unnamed protein product [Mus musculus]
Length = 111
Score = 65.9 bits (159), Expect = 5e-10
Identities = 33/74 (44%), Positives = 42/74 (56%)
Frame = -3
Query: 683 VYLGYHTLAQVFAGTILGVSLGAIWFWVVNTVLRPYFSLIEESEFGKRFYVKDTSHIPNV 504
VYL YHT +QVF G + G S+ WF + +L P F I + F ++DTS IPNV
Sbjct: 28 VYLLYHTWSQVFYGGVAGSSMAVAWFIITQEILTPLFPRIAAWPISEFFLIRDTSLIPNV 87
Query: 503 LKFEYEQARAARKN 462
L FEY RA +N
Sbjct: 88 LWFEYTVTRAEARN 101
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 564,141,401
Number of Sequences: 1393205
Number of extensions: 11895953
Number of successful extensions: 31442
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 30546
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31433
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)