Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004581A_C01 KMC004581A_c01
(688 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
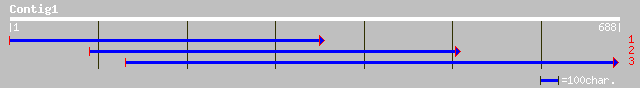
ref|NP_178213.1| hypothetical protein; protein id: At1g80980.1 [... 54 2e-06
ref|NP_178185.1| hypothetical protein; protein id: At1g80700.1, ... 54 2e-06
emb|CAD59239.1| NAD(+) ADP-ribosyltransferase-3 [Dictyostelium d... 50 3e-05
ref|NP_500151.1| Putative prenylated nuclear protein, with 2 coi... 47 3e-04
emb|CAD21337.1| conserved hypothetical protein [Neurospora crass... 44 0.002
>ref|NP_178213.1| hypothetical protein; protein id: At1g80980.1 [Arabidopsis
thaliana] gi|25372794|pir||H96842 F23A5.34 [imported] -
Arabidopsis thaliana
gi|6503310|gb|AAF14686.1|AC011713_34 F23A5.34
[Arabidopsis thaliana]
Length = 214
Score = 54.3 bits (129), Expect = 2e-06
Identities = 29/61 (47%), Positives = 40/61 (65%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSSSNVAKN 507
+EEE+KEKE+ + E KS +LME++ RL K+EE +KEIV+ETKK S + K
Sbjct: 123 KEEEKKEKEEAEQKALQVEAATKSHEELMEMRQRLGKIEETIKEIVLETKKPSGNAPTKT 182
Query: 506 Q 504
Q
Sbjct: 183 Q 183
>ref|NP_178185.1| hypothetical protein; protein id: At1g80700.1, supported by cDNA:
gi_20466575 [Arabidopsis thaliana]
gi|25372793|pir||D96839 F23A5.5 [imported] - Arabidopsis
thaliana gi|6503283|gb|AAF14659.1|AC011713_7 F23A5.5
[Arabidopsis thaliana] gi|20466576|gb|AAM20605.1|
unknown protein [Arabidopsis thaliana]
gi|22136386|gb|AAM91271.1| unknown protein [Arabidopsis
thaliana]
Length = 214
Score = 54.3 bits (129), Expect = 2e-06
Identities = 29/61 (47%), Positives = 40/61 (65%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSSSNVAKN 507
+EEE+KEKE+ + E KS +LME++ RL K+EE +KEIV+ETKK S + K
Sbjct: 123 KEEEKKEKEEAEQKALQVEAATKSHEELMEMRQRLGKIEETIKEIVLETKKPSGNAPTKT 182
Query: 506 Q 504
Q
Sbjct: 183 Q 183
>emb|CAD59239.1| NAD(+) ADP-ribosyltransferase-3 [Dictyostelium discoideum]
Length = 2536
Score = 50.1 bits (118), Expect = 3e-05
Identities = 35/135 (25%), Positives = 67/135 (48%), Gaps = 4/135 (2%)
Frame = -3
Query: 686 EEEEQKEKEKE--MELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSSSNVA 513
++E++KEKEKE + E+ + K+ Q + K + +KL+E KE + K S
Sbjct: 176 QKEKEKEKEKEKAKKFKEKEKEKEKAKKQKEKEKEKAKKLKEKEKE--KKKKNSESEKPK 233
Query: 512 KNQTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGEESKGSMATPNS 333
K+ + ++ +N S S + +S+ +++ SK +S K K E+SK + +
Sbjct: 234 KSNKSKKQEDTENESESESESESESSSSESEKEESSKKSSSKKKTTTKEKSKPTKKSKKQ 293
Query: 332 SLQDPK--GQNQSKG 294
+D + G ++S G
Sbjct: 294 ETEDEEESGSDESSG 308
Score = 44.3 bits (103), Expect = 0.002
Identities = 40/153 (26%), Positives = 70/153 (45%), Gaps = 20/153 (13%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVV---ETKKQSSSNV 516
E+E++KEKEK + E+ + K+ Q + K + +KL+E KE E++K SN
Sbjct: 178 EKEKEKEKEKAKKFKEKEKEKEKAKKQKEKEKEKAKKLKEKEKEKKKKNSESEKPKKSNK 237
Query: 515 AKNQT---------ADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGE- 366
+K Q ++ E +S + ++ SS +S++ + + KSK E
Sbjct: 238 SKKQEDTENESESESESESESSSSESEKEESSKKSSSKKKTTTKEKSKPTKKSKKQETED 297
Query: 365 -------ESKGSMATPNSSLQDPKGQNQSKGAS 288
ES G +SS +D + + SK +S
Sbjct: 298 EEESGSDESSGDDDEKSSSDEDEREKKSSKKSS 330
>ref|NP_500151.1| Putative prenylated nuclear protein, with 2 coiled coil-4 domains,
nematode specific [Caenorhabditis elegans]
gi|7105679|gb|AAF36065.1| Hypothetical protein Y76B12C.4
[Caenorhabditis elegans]
Length = 252
Score = 46.6 bits (109), Expect = 3e-04
Identities = 36/112 (32%), Positives = 55/112 (48%), Gaps = 2/112 (1%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSSSNVAKN 507
+E +KEKEKE E E+ ++D + E + EK E+ KE+ E +K+ K
Sbjct: 67 KEAAEKEKEKEKEKEKTHEKN-EADKEEKEAEKEEEKEEKPPKEVTEEKEKEKERE--KT 123
Query: 506 QTADDGKELQNSSAPRDTSSASTSNRAVDEDR--FSKHNSLKSKPNLGEESK 357
+T D+ +E APR+ + ++ DED SK LK K L +E K
Sbjct: 124 KTKDELEE-----APREKETMEKKQKSKDEDEGDLSKEEILKIKKALKKEKK 170
Score = 35.8 bits (81), Expect = 0.57
Identities = 32/118 (27%), Positives = 54/118 (45%), Gaps = 5/118 (4%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSS-----S 522
EE+E +++E++ E P E E K + E ++LEEA +E KKQ S
Sbjct: 92 EEKEAEKEEEKEEKPPKEVTEEKEKEKEREKTKTKDELEEAPREKETMEKKQKSKDEDEG 151
Query: 521 NVAKNQTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGEESKGSM 348
+++K + K L+ RD T +D+ +K+++ S E+ K SM
Sbjct: 152 DLSKEEILKIKKALKKEKKLRDRERLKTEK--IDKSTETKNSTTPST----EKEKSSM 203
Score = 34.3 bits (77), Expect = 1.7
Identities = 26/101 (25%), Positives = 43/101 (41%), Gaps = 11/101 (10%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEE--REAKSDPQLMEVKVRLEKLEEA---------LKEIVVET 540
E E+KEKEKE E ++ EA + + ME K + + +E +K+ + +
Sbjct: 109 EVTEEKEKEKEREKTKTKDELEEAPREKETMEKKQKSKDEDEGDLSKEEILKIKKALKKE 168
Query: 539 KKQSSSNVAKNQTADDGKELQNSSAPRDTSSASTSNRAVDE 417
KK K + D E +NS+ P S+ + E
Sbjct: 169 KKLRDRERLKTEKIDKSTETKNSTTPSTEKEKSSMKKTKKE 209
>emb|CAD21337.1| conserved hypothetical protein [Neurospora crassa]
gi|28923265|gb|EAA32481.1| predicted protein [Neurospora
crassa]
Length = 1750
Score = 44.3 bits (103), Expect = 0.002
Identities = 40/141 (28%), Positives = 65/141 (45%), Gaps = 20/141 (14%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLE-------------EALKEIVV 546
E+E++ EKE+E E +ERE + + +L + + R ++LE E KE
Sbjct: 1385 EKEKELEKEREREKELEKERELEKEKELEKEREREKELEKEREKEKERELEKEKEKEKEK 1444
Query: 545 ETKKQSSSNVAKNQTADD------GKELQNSS-APRDTSSASTSNRAVDEDRFSKHNSLK 387
E ++Q +SN K ADD G+ + S P D T N A+ ++ K
Sbjct: 1445 EKEQQGASNSLKRGAADDDDDEPSGRSSKRSKHTPEDN---PTQNPALSPNKKRKTPPPD 1501
Query: 386 SKPNLGEESKGSMATPNSSLQ 324
++P + +KG AT +S Q
Sbjct: 1502 TEPKPAKRAKGEPATRSSERQ 1522
Score = 41.6 bits (96), Expect = 0.010
Identities = 39/156 (25%), Positives = 63/156 (40%), Gaps = 25/156 (16%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQL---------------MEVKVRLEKLEEALKEI 552
E E +KEKE E E +ERE + + +L +E + LEK E KE+
Sbjct: 1363 ERELEKEKELEKERELEKERELEKEKELEKEREREKELEKERELEKEKELEKEREREKEL 1422
Query: 551 VVETKKQSSSNVAKNQTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSK--- 381
E +K+ + K + + KE + A + + D+D S +S +SK
Sbjct: 1423 EKEREKEKERELEKEKEKEKEKEKEQQGASNSLKRGAADD---DDDEPSGRSSKRSKHTP 1479
Query: 380 -------PNLGEESKGSMATPNSSLQDPKGQNQSKG 294
P L K P++ +PK ++KG
Sbjct: 1480 EDNPTQNPALSPNKKRKTPPPDT---EPKPAKRAKG 1512
Score = 40.8 bits (94), Expect = 0.018
Identities = 33/134 (24%), Positives = 59/134 (43%), Gaps = 4/134 (2%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSSSNVAKN 507
E E +KE+E E E +ERE + + +L E + LEK E KE +E +++ + K
Sbjct: 1345 ERELEKERELEKERELEKERELEKEKEL-EKERELEKERELEKEKELEKEREREKELEKE 1403
Query: 506 QTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGEESKGSMATPNSSL 327
+ + KEL+ R+ R +++R + K K E+ S + +
Sbjct: 1404 RELEKEKELEKE---REREKELEKEREKEKERELEKEKEKEKEKEKEQQGASNSLKRGAA 1460
Query: 326 QD----PKGQNQSK 297
D P G++ +
Sbjct: 1461 DDDDDEPSGRSSKR 1474
Score = 37.0 bits (84), Expect = 0.26
Identities = 30/118 (25%), Positives = 51/118 (42%), Gaps = 10/118 (8%)
Frame = -3
Query: 686 EEEEQKEKEKE---MELNPPEEREAKSDPQL-----MEVKVRLEKLEEALKEIVVETKK- 534
EE+EQK+KE++ EL +ERE + D +L +E + LEK E KE +E ++
Sbjct: 1216 EEQEQKQKEEQAQAQELEKEKERELEKDRELEKDRELEKERELEKERELEKERELEKERE 1275
Query: 533 -QSSSNVAKNQTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGEE 363
+ + K + + +EL+ E K L+ + L +E
Sbjct: 1276 LEKERELEKERELEKERELEKERELEKEKELEKERELEKERELEKERELEKERELEKE 1333
Score = 33.9 bits (76), Expect = 2.2
Identities = 28/110 (25%), Positives = 46/110 (41%), Gaps = 2/110 (1%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKK--QSSSNVA 513
E E +KE+E E E +ERE + + +L E + LEK E KE +E +K + +
Sbjct: 1255 ERELEKERELEKERELEKERELEKEREL-EKERELEKERELEKERELEKEKELEKERELE 1313
Query: 512 KNQTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGEE 363
K + + +EL+ E K L+ + L +E
Sbjct: 1314 KERELEKERELEKERELEKERELEKERELEKERELEKERELEKERELEKE 1363
Score = 33.1 bits (74), Expect = 3.7
Identities = 27/108 (25%), Positives = 46/108 (42%)
Frame = -3
Query: 686 EEEEQKEKEKEMELNPPEEREAKSDPQLMEVKVRLEKLEEALKEIVVETKKQSSSNVAKN 507
E E +KE+E E E +ERE + + +L E + LEK E KE +E +++ + K
Sbjct: 1273 ERELEKERELEKERELEKERELEKEREL-EKEKELEKERELEKERELEKERE----LEKE 1327
Query: 506 QTADDGKELQNSSAPRDTSSASTSNRAVDEDRFSKHNSLKSKPNLGEE 363
+ + +EL+ E K L+ + L +E
Sbjct: 1328 RELEKERELEKERELEKERELEKERELEKERELEKERELEKEKELEKE 1375
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,428,263
Number of Sequences: 1393205
Number of extensions: 11611305
Number of successful extensions: 62060
Number of sequences better than 10.0: 897
Number of HSP's better than 10.0 without gapping: 43060
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56148
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30835865868
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)