Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004572A_C01 KMC004572A_c01
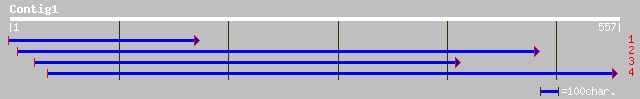
(554 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB91833.1| putative ribosomal RNA apurinic site specific ly... 34 1.0
ref|NP_438234.1| recombination protein (rec2) [Haemophilus influ... 33 1.8
gb|AAC13733.1| recombination protein 33 1.8
gb|AAD02491.1| unknown [Fiji disease virus] 33 3.0
ref|NP_190517.1| putative protein; protein id: At3g49480.1 [Arab... 32 6.8
>dbj|BAB91833.1| putative ribosomal RNA apurinic site specific lyase [Oryza sativa
(japonica cultivar-group)]
Length = 510
Score = 34.3 bits (77), Expect = 1.0
Identities = 14/34 (41%), Positives = 22/34 (64%)
Frame = -3
Query: 465 LTFIHYQGYQGFTDELEFLAYVLREGIVLERMII 364
+ ++ + GY+G EL FL Y+L G VL+ M+I
Sbjct: 412 IKYVVFHGYRGDRSELTFLKYILGSGQVLQEMVI 445
>ref|NP_438234.1| recombination protein (rec2) [Haemophilus influenzae Rd]
gi|1172877|sp|P44408|REC2_HAEIN Recombination protein 2
gi|1075204|pir||I64045 recombination protein rec2 -
Haemophilus influenzae (strain Rd KW20)
gi|1573009|gb|AAC21739.1| recombination protein (rec2)
[Haemophilus influenzae Rd]
Length = 788
Score = 33.5 bits (75), Expect = 1.8
Identities = 16/27 (59%), Positives = 20/27 (73%)
Frame = -3
Query: 435 GFTDELEFLAYVLREGIVLERMIISTD 355
G ELE L Y+ REGIVLE++I+S D
Sbjct: 568 GSMAELEILPYLQREGIVLEKLILSHD 594
>gb|AAC13733.1| recombination protein
Length = 800
Score = 33.5 bits (75), Expect = 1.8
Identities = 16/27 (59%), Positives = 20/27 (73%)
Frame = -3
Query: 435 GFTDELEFLAYVLREGIVLERMIISTD 355
G ELE L Y+ REGIVLE++I+S D
Sbjct: 568 GSMAELEILPYLQREGIVLEKLILSHD 594
>gb|AAD02491.1| unknown [Fiji disease virus]
Length = 1146
Score = 32.7 bits (73), Expect = 3.0
Identities = 16/30 (53%), Positives = 20/30 (66%)
Frame = +3
Query: 24 CLKLKSQDKYKISPINPKTQTDNIQQRQYN 113
CLKL +Y IS +N KTQ +NI R+YN
Sbjct: 153 CLKLNVPIEYYISMLNLKTQDNNIFLREYN 182
>ref|NP_190517.1| putative protein; protein id: At3g49480.1 [Arabidopsis thaliana]
gi|11358455|pir||T46226 hypothetical protein T9C5.80 -
Arabidopsis thaliana gi|6561949|emb|CAB62453.1| putative
protein [Arabidopsis thaliana]
Length = 309
Score = 31.6 bits (70), Expect = 6.8
Identities = 17/56 (30%), Positives = 26/56 (46%)
Frame = -3
Query: 528 NNEPSTWGKPPRAPIFHHVLHLTFIHYQGYQGFTDELEFLAYVLREGIVLERMIIS 361
N++P+ W +P P HL + GY G DE + + Y+L L+ IS
Sbjct: 218 NHDPALWNQPSSIPRCLSS-HLEIFEWDGYVGREDEKKIIRYILENSKYLKTAGIS 272
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 414,499,324
Number of Sequences: 1393205
Number of extensions: 7739482
Number of successful extensions: 16982
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 16623
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16981
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)