Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004564A_C01 KMC004564A_c01
(606 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
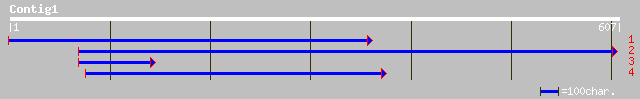
ref|NP_566423.1| expressed protein; protein id: At3g12400.1 [Ara... 79 5e-14
ref|NP_196890.1| putative protein; protein id: At5g13860.1 [Arab... 76 4e-13
ref|XP_214938.1| similar to tumor susceptibility gene 101 protei... 36 0.34
gb|AAF87776.1| tumor susceptibility protein 101 [Chelonia mydas] 36 0.34
ref|NP_006283.1| tumor susceptibility gene 101 [Homo sapiens] gi... 36 0.34
>ref|NP_566423.1| expressed protein; protein id: At3g12400.1 [Arabidopsis thaliana]
gi|12321968|gb|AAG51025.1|AC069474_24 unknown protein;
81998-83194 [Arabidopsis thaliana]
gi|15795159|dbj|BAB03147.1| mouse and human tumor
susceptibility gene-like protein [Arabidopsis thaliana]
gi|15810489|gb|AAL07132.1| unknown protein [Arabidopsis
thaliana] gi|21593369|gb|AAM65318.1| unknown
[Arabidopsis thaliana] gi|22136968|gb|AAM91713.1|
unknown protein [Arabidopsis thaliana]
Length = 398
Score = 79.0 bits (193), Expect = 5e-14
Identities = 37/57 (64%), Positives = 47/57 (81%)
Frame = -1
Query: 606 DTLYALDKAVQVGAVPFDQYLRSVRALSREQFFQRATAAKVKAAQMQAQVSNMAARI 436
D +Y+LDK+ Q G VPFDQYLR+VR LSREQFF RAT +KV+AAQM+ QV+ +A R+
Sbjct: 340 DAIYSLDKSFQDGVVPFDQYLRNVRLLSREQFFHRATGSKVRAAQMEVQVAAIAGRL 396
>ref|NP_196890.1| putative protein; protein id: At5g13860.1 [Arabidopsis thaliana]
gi|10177652|dbj|BAB11114.1| contains similarity to
nascent polypeptide associated complex alpha
chain~gene_id:MAC12.18 [Arabidopsis thaliana]
Length = 368
Score = 75.9 bits (185), Expect = 4e-13
Identities = 35/57 (61%), Positives = 48/57 (83%)
Frame = -1
Query: 606 DTLYALDKAVQVGAVPFDQYLRSVRALSREQFFQRATAAKVKAAQMQAQVSNMAARI 436
D +Y++DK+ + G++PFDQYLR+VR LSREQFF RATA KV+ QM AQV+++AAR+
Sbjct: 310 DVVYSMDKSFRDGSLPFDQYLRNVRLLSREQFFHRATAEKVREIQMDAQVASIAARL 366
>ref|XP_214938.1| similar to tumor susceptibility gene 101 protein [Mus musculus]
[Rattus norvegicus]
Length = 149
Score = 36.2 bits (82), Expect = 0.34
Identities = 17/42 (40%), Positives = 27/42 (63%)
Frame = -1
Query: 606 DTLYALDKAVQVGAVPFDQYLRSVRALSREQFFQRATAAKVK 481
DT++ L +A++ G + D +L+ VR LSR+QF RA K +
Sbjct: 99 DTIFYLGEALRRGVIDLDVFLKHVRLLSRKQFQLRALMQKAR 140
>gb|AAF87776.1| tumor susceptibility protein 101 [Chelonia mydas]
Length = 392
Score = 36.2 bits (82), Expect = 0.34
Identities = 17/42 (40%), Positives = 27/42 (63%)
Frame = -1
Query: 606 DTLYALDKAVQVGAVPFDQYLRSVRALSREQFFQRATAAKVK 481
DT++ L +A++ G + D +L+ VR LSR+QF RA K +
Sbjct: 342 DTIFYLGEALRRGVIDLDVFLKHVRLLSRKQFQLRALMQKAR 383
>ref|NP_006283.1| tumor susceptibility gene 101 [Homo sapiens]
gi|9789790|sp|Q99816|T101_HUMAN Tumor susceptibility
gene 101 protein gi|3184258|gb|AAC52083.1| tumor
susceptibility protein [Homo sapiens]
Length = 390
Score = 36.2 bits (82), Expect = 0.34
Identities = 17/42 (40%), Positives = 27/42 (63%)
Frame = -1
Query: 606 DTLYALDKAVQVGAVPFDQYLRSVRALSREQFFQRATAAKVK 481
DT++ L +A++ G + D +L+ VR LSR+QF RA K +
Sbjct: 340 DTIFYLGEALRRGVIDLDVFLKHVRLLSRKQFQLRALMQKAR 381
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,743,923
Number of Sequences: 1393205
Number of extensions: 10338447
Number of successful extensions: 26849
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 25921
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26832
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)