Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004560A_C01 KMC004560A_c01
(562 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
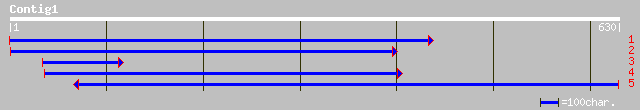
ref|NP_195981.1| putative protein; protein id: At5g03610.1, supp... 142 3e-33
gb|AAO41990.1| putative lipase acylhydrolase [Arabidopsis thaliana] 131 5e-30
ref|NP_187604.1| putative lipase acylhydrolase; protein id: At3g... 130 8e-30
gb|AAF04430.1|AC010927_23 unknown protein [Arabidopsis thaliana] 130 8e-30
ref|NP_195980.1| putative protein; protein id: At5g03600.1 [Arab... 105 3e-22
>ref|NP_195981.1| putative protein; protein id: At5g03610.1, supported by cDNA:
13022., supported by cDNA: gi_13430833 [Arabidopsis
thaliana] gi|11357496|pir||T48388 hypothetical protein
F17C15.30 - Arabidopsis thaliana
gi|7340646|emb|CAB82926.1| putative protein [Arabidopsis
thaliana] gi|13430834|gb|AAK26039.1|AF360329_1 unknown
protein [Arabidopsis thaliana]
gi|21537326|gb|AAM61667.1| putative lipase/acylhydrolase
[Arabidopsis thaliana]
Length = 359
Score = 142 bits (358), Expect = 3e-33
Identities = 67/166 (40%), Positives = 107/166 (64%), Gaps = 1/166 (0%)
Frame = -3
Query: 554 SFTESLIKQLTLNLKRVQSLGIGKVAVGLLQPVGCLPGLTVVSFHLSCIDPLNLVSKTHN 375
+F + ++ Q +NL+R+ +LG+ K+AV LQP+GCLP T V+ C + N + HN
Sbjct: 188 AFIKQVVDQTEVNLRRIHALGVKKIAVPSLQPLGCLPPFTFVTSFQRCNETQNALVNLHN 247
Query: 374 QMLLQTVQELNKEMGKPVFITLDLYNSFLTTIKTMQKKHAENSTLMNPLQPCCEGINFK- 198
+L Q V +LN E + FI LDLYN+FLT K + + ++ +PL+PCC G++ +
Sbjct: 248 NLLQQVVAKLNNETKQSTFIILDLYNAFLTVFKN-KGSNPGSTRFESPLKPCCVGVSREY 306
Query: 197 SCGSVNDKGEKQYKLCKKPEFSLFWDNVHPSQNGWHAVSMLLQPSL 60
+CGSV++KG K+Y +C P+ + FWD +HP++ GW +V +L+ SL
Sbjct: 307 NCGSVDEKGVKKYIVCDNPKTAFFWDGLHPTEEGWRSVYSVLRESL 352
>gb|AAO41990.1| putative lipase acylhydrolase [Arabidopsis thaliana]
Length = 354
Score = 131 bits (330), Expect = 5e-30
Identities = 61/168 (36%), Positives = 97/168 (57%), Gaps = 1/168 (0%)
Frame = -3
Query: 560 IKSFTESLIKQLTLNLKRVQSLGIGKVAVGLLQPVGCLPGLTVVSFHLSCIDPLNLVSKT 381
+ +F + ++ Q+ +N R+ LG+ K+ + +QP+GCLP +TV + C N +
Sbjct: 183 LPAFMKQVVDQIAVNAMRIHKLGVNKIVIPSMQPLGCLPSITVFNSFQRCNATDNASTNL 242
Query: 380 HNQMLLQTVQELNKEMGKPVFITLDLYNSFLTTIKTMQKKHAENSTLMNPLQPCCEGINF 201
HN +L + + LN E + F+ LD YN+FLT K + S NPL+PCC G+N
Sbjct: 243 HNYLLHKAIARLNNETKQSTFVVLDHYNAFLTVFKNKGPEPGV-SRFGNPLKPCCVGVNS 301
Query: 200 K-SCGSVNDKGEKQYKLCKKPEFSLFWDNVHPSQNGWHAVSMLLQPSL 60
C +V++KGEK+Y +C+ P+ + FWD HPS+ GW +V +L L
Sbjct: 302 SYDCSNVDEKGEKKYIICEDPKAAFFWDIFHPSEEGWRSVYSVLHKHL 349
>ref|NP_187604.1| putative lipase acylhydrolase; protein id: At3g09930.1 [Arabidopsis
thaliana] gi|6681326|gb|AAF23243.1|AC015985_1 putative
lipase/acylhydrolase [Arabidopsis thaliana]
Length = 354
Score = 130 bits (328), Expect = 8e-30
Identities = 61/168 (36%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Frame = -3
Query: 560 IKSFTESLIKQLTLNLKRVQSLGIGKVAVGLLQPVGCLPGLTVVSFHLSCIDPLNLVSKT 381
+ +F + ++ Q+ +N R+ LG+ K+ + +QP+GCLP +TV + C N +
Sbjct: 183 LPAFMKQVVDQIAVNAMRIHKLGVNKIVIPSMQPLGCLPSITVFNSFQRCNATDNASTNL 242
Query: 380 HNQMLLQTVQELNKEMGKPVFITLDLYNSFLTTIKTMQKKHAENSTLMNPLQPCCEGINF 201
HN +L + + LN E F+ LD YN+FLT K + S NPL+PCC G+N
Sbjct: 243 HNYLLHKAIARLNNETKPSTFVVLDHYNAFLTVFKNKGPEPGV-SRFGNPLKPCCVGVNS 301
Query: 200 K-SCGSVNDKGEKQYKLCKKPEFSLFWDNVHPSQNGWHAVSMLLQPSL 60
C +V++KGEK+Y +C+ P+ + FWD HPS+ GW +V +L L
Sbjct: 302 SYDCSNVDEKGEKKYIICEDPKAAFFWDIFHPSEEGWRSVYSVLHKHL 349
>gb|AAF04430.1|AC010927_23 unknown protein [Arabidopsis thaliana]
Length = 270
Score = 130 bits (328), Expect = 8e-30
Identities = 61/168 (36%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Frame = -3
Query: 560 IKSFTESLIKQLTLNLKRVQSLGIGKVAVGLLQPVGCLPGLTVVSFHLSCIDPLNLVSKT 381
+ +F + ++ Q+ +N R+ LG+ K+ + +QP+GCLP +TV + C N +
Sbjct: 99 LPAFMKQVVDQIAVNAMRIHKLGVNKIVIPSMQPLGCLPSITVFNSFQRCNATDNASTNL 158
Query: 380 HNQMLLQTVQELNKEMGKPVFITLDLYNSFLTTIKTMQKKHAENSTLMNPLQPCCEGINF 201
HN +L + + LN E F+ LD YN+FLT K + S NPL+PCC G+N
Sbjct: 159 HNYLLHKAIARLNNETKPSTFVVLDHYNAFLTVFKNKGPEPGV-SRFGNPLKPCCVGVNS 217
Query: 200 K-SCGSVNDKGEKQYKLCKKPEFSLFWDNVHPSQNGWHAVSMLLQPSL 60
C +V++KGEK+Y +C+ P+ + FWD HPS+ GW +V +L L
Sbjct: 218 SYDCSNVDEKGEKKYIICEDPKAAFFWDIFHPSEEGWRSVYSVLHKHL 265
>ref|NP_195980.1| putative protein; protein id: At5g03600.1 [Arabidopsis thaliana]
gi|11357495|pir||T48387 hypothetical protein F17C15.20 -
Arabidopsis thaliana gi|7340645|emb|CAB82925.1| putative
protein [Arabidopsis thaliana]
Length = 322
Score = 105 bits (263), Expect = 3e-22
Identities = 61/175 (34%), Positives = 95/175 (53%), Gaps = 5/175 (2%)
Frame = -3
Query: 560 IKSFTESLIKQLTLNLKRVQSLGIGKVAVGLLQPVGCLPGLTVVSFHLSCIDPLNLVSKT 381
+K+ E ++ L +N+ + L K+AV LQP+GCLP T S SC + + + +
Sbjct: 155 LKALVEKVVDNLRVNMIVLGGLLFKKIAVTSLQPIGCLPSYTSASSFKSCNESQSALVEL 214
Query: 380 HNQMLLQTVQELNKE----MGKPVFITLDLYNSFLTTIKTMQKKHAENSTLMNPLQPCCE 213
HN++L + V +LN++ + F +D++N+F+T +K K NP++ CCE
Sbjct: 215 HNKLLKKVVAKLNEQSRVMKKEQHFFIIDIHNAFMTVMKNKGSKR-----FKNPMKSCCE 269
Query: 212 GINFKSCGSVNDKGEKQYKLCKKPEFSLFWDNVHPSQNGWHAV-SMLLQPSLDQL 51
G CG +D G K Y LC P+ FWD VHP+Q GW ++ S+L P D L
Sbjct: 270 GY----CGRSSD-GGKLYTLCDDPKSFFFWDAVHPTQEGWRSIYSVLGNPLTDFL 319
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,185,959
Number of Sequences: 1393205
Number of extensions: 10016370
Number of successful extensions: 35692
Number of sequences better than 10.0: 204
Number of HSP's better than 10.0 without gapping: 34498
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35480
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)