Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004502A_C02 KMC004502A_c02
(631 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
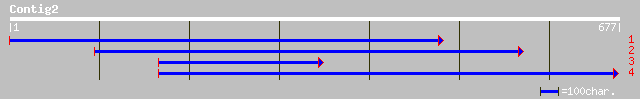
ref|NP_193986.1| putative protein; protein id: At4g22550.1 [Arab... 62 8e-09
gb|AAG49895.1| PnFL-1 [Ipomoea nil] 56 4e-07
ref|NP_723543.1| CG31717-PA [Drosophila melanogaster] gi|1844710... 52 8e-06
pir||T16869 hypothetical protein T13C5.6 - Caenorhabditis elegans 41 0.011
ref|NP_509040.2| PAP2 superfamily [Caenorhabditis elegans] gi|20... 41 0.011
>ref|NP_193986.1| putative protein; protein id: At4g22550.1 [Arabidopsis thaliana]
gi|7486605|pir||T05449 hypothetical protein F7K2.130 -
Arabidopsis thaliana gi|3892710|emb|CAA22160.1| putative
protein [Arabidopsis thaliana]
gi|7269101|emb|CAB79210.1| putative protein [Arabidopsis
thaliana] gi|28973659|gb|AAO64149.1| unknown protein
[Arabidopsis thaliana]
Length = 213
Score = 61.6 bits (148), Expect = 8e-09
Identities = 27/53 (50%), Positives = 37/53 (68%)
Frame = -1
Query: 613 LVAPVWSWALTTVVSRVVLGRHYILDVLFGACFGVLEALITLRLLQYRALI*G 455
+V VW WA T +SR++LGRHY+LDV GA G++EAL LR L++ +I G
Sbjct: 160 VVVVVWIWATVTAISRILLGRHYVLDVAAGAFLGIVEALFALRFLRFDEMIFG 212
>gb|AAG49895.1| PnFL-1 [Ipomoea nil]
Length = 209
Score = 55.8 bits (133), Expect = 4e-07
Identities = 24/46 (52%), Positives = 32/46 (69%)
Frame = -1
Query: 601 VWSWALTTVVSRVVLGRHYILDVLFGACFGVLEALITLRLLQYRAL 464
+ +WA T VSR++LGRH++LDV+ GAC GVLE L + R Y L
Sbjct: 159 IGAWATITSVSRILLGRHFVLDVVAGACLGVLEGLFSFRFFNYDKL 204
>ref|NP_723543.1| CG31717-PA [Drosophila melanogaster] gi|18447108|gb|AAL68145.1|
AT30094p [Drosophila melanogaster]
gi|22946111|gb|AAN10729.1| CG31717-PA [Drosophila
melanogaster]
Length = 204
Score = 51.6 bits (122), Expect = 8e-06
Identities = 22/41 (53%), Positives = 31/41 (74%)
Frame = -1
Query: 616 LLVAPVWSWALTTVVSRVVLGRHYILDVLFGACFGVLEALI 494
+ + PV +WA++ +SR++L RHYILD+ GA GVLEALI
Sbjct: 137 IFLMPVTAWAVSVAISRLILQRHYILDICAGAAIGVLEALI 177
>pir||T16869 hypothetical protein T13C5.6 - Caenorhabditis elegans
Length = 345
Score = 41.2 bits (95), Expect = 0.011
Identities = 23/48 (47%), Positives = 27/48 (55%)
Frame = -1
Query: 625 AVNLLVAPVWSWALTTVVSRVVLGRHYILDVLFGACFGVLEALITLRL 482
A L V P + L +SRV LGRHYI DVL G G LEA + L +
Sbjct: 126 AAPLYVIPFIPFPLVVGLSRVALGRHYITDVLAGIFIGYLEARLMLTI 173
>ref|NP_509040.2| PAP2 superfamily [Caenorhabditis elegans]
gi|20198885|gb|AAM15605.1|U39648_8 Hypothetical protein
T13C5.6 [Caenorhabditis elegans]
Length = 186
Score = 41.2 bits (95), Expect = 0.011
Identities = 23/48 (47%), Positives = 27/48 (55%)
Frame = -1
Query: 625 AVNLLVAPVWSWALTTVVSRVVLGRHYILDVLFGACFGVLEALITLRL 482
A L V P + L +SRV LGRHYI DVL G G LEA + L +
Sbjct: 126 AAPLYVIPFIPFPLVVGLSRVALGRHYITDVLAGIFIGYLEARLMLTI 173
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 523,382,432
Number of Sequences: 1393205
Number of extensions: 11060767
Number of successful extensions: 34426
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 31685
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34043
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)