Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
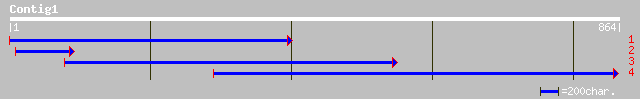
Query= KMC004488A_C01 KMC004488A_c01
(864 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||A86229 hypothetical protein [imported] - Arabidopsis thalia... 47 3e-04
pir||T51668 myb-related transcription factor MYB61 [imported] - ... 47 5e-04
ref|NP_172425.2| myb family transcription factor; protein id: At... 47 5e-04
gb|AAO49411.1|AF485893_1 MYB2 [Dendrobium sp. XMW-2002-2] 41 0.026
dbj|BAB39972.1| putative transcription factor (myb) [Oryza sativ... 40 0.059
>pir||A86229 hypothetical protein [imported] - Arabidopsis thaliana
gi|3482929|gb|AAC33214.1| Putative transcription factor
[Arabidopsis thaliana]
Length = 414
Score = 47.4 bits (111), Expect = 3e-04
Identities = 34/125 (27%), Positives = 59/125 (47%), Gaps = 11/125 (8%)
Frame = -1
Query: 858 VNGSNYWEASASN---NSNSSNTSDGIGSRELSSSNILSSSSWGLTDCSSSTEEAKWCEY 688
++G ++ + +A N +N++NTS+ + S S SS+S + + EE KW EY
Sbjct: 239 ISGGDHVKLAAPNWEFQTNNNNTSNFFDNGGFSWSIPNSSTSSSQVKPNHNFEEIKWSEY 298
Query: 687 LQNPMLMLEAAQNQTPSKHLVPDTLGAMLPHSKQQEEPSQ--------SSSIFSKDLQKL 532
L P + Q+QT + + S + SQ +S +FSKDLQ++
Sbjct: 299 LNTPFFIGSTVQSQTSQPIYIKSETDYLANVSNMTDPWSQNENLGTTETSDVFSKDLQRM 358
Query: 531 TAAFE 517
+F+
Sbjct: 359 AVSFD 363
>pir||T51668 myb-related transcription factor MYB61 [imported] - Arabidopsis
thaliana (fragment) gi|3941484|gb|AAC83618.1| putative
transcription factor [Arabidopsis thaliana]
Length = 291
Score = 46.6 bits (109), Expect = 5e-04
Identities = 34/124 (27%), Positives = 58/124 (46%), Gaps = 11/124 (8%)
Frame = -1
Query: 858 VNGSNYWEASASN---NSNSSNTSDGIGSRELSSSNILSSSSWGLTDCSSSTEEAKWCEY 688
++G ++ + +A N +N++NTS+ + S S SS+S + + EE KW EY
Sbjct: 164 ISGGDHVKLAAPNWEFQTNNNNTSNFFDNGGFSWSIPNSSTSSSQVKPNHNFEEIKWSEY 223
Query: 687 LQNPMLMLEAAQNQTPSKHLVPDTLGAMLPHSKQQEEPSQ--------SSSIFSKDLQKL 532
L P + Q+QT + + S + SQ +S +FSKDLQ++
Sbjct: 224 LNTPFFIGSTVQSQTSQPIYIKSETDYLANVSNMTDPWSQNENLGTTETSDVFSKDLQRM 283
Query: 531 TAAF 520
+F
Sbjct: 284 AVSF 287
>ref|NP_172425.2| myb family transcription factor; protein id: At1g09540.1, supported
by cDNA: gi_20465974 [Arabidopsis thaliana]
gi|17380966|gb|AAL36295.1| putative transcription factor
[Arabidopsis thaliana] gi|20465975|gb|AAM20173.1|
putative transcription factor protein [Arabidopsis
thaliana]
Length = 366
Score = 46.6 bits (109), Expect = 5e-04
Identities = 34/124 (27%), Positives = 58/124 (46%), Gaps = 11/124 (8%)
Frame = -1
Query: 858 VNGSNYWEASASN---NSNSSNTSDGIGSRELSSSNILSSSSWGLTDCSSSTEEAKWCEY 688
++G ++ + +A N +N++NTS+ + S S SS+S + + EE KW EY
Sbjct: 239 ISGGDHVKLAAPNWEFQTNNNNTSNFFDNGGFSWSIPNSSTSSSQVKPNHNFEEIKWSEY 298
Query: 687 LQNPMLMLEAAQNQTPSKHLVPDTLGAMLPHSKQQEEPSQ--------SSSIFSKDLQKL 532
L P + Q+QT + + S + SQ +S +FSKDLQ++
Sbjct: 299 LNTPFFIGSTVQSQTSQPIYIKSETDYLANVSNMTDPWSQNENLGTTETSDVFSKDLQRM 358
Query: 531 TAAF 520
+F
Sbjct: 359 AVSF 362
>gb|AAO49411.1|AF485893_1 MYB2 [Dendrobium sp. XMW-2002-2]
Length = 371
Score = 40.8 bits (94), Expect = 0.026
Identities = 31/113 (27%), Positives = 50/113 (43%), Gaps = 10/113 (8%)
Frame = -1
Query: 843 YWEA-SASNNSNSSNTSDGIGSRELSSSNILSSSSWGLTDCSSSTEEAKWCEYLQNPMLM 667
YWE ++SN+S SS ++D + + L TE+ KW EYL + +
Sbjct: 255 YWETGNSSNSSASSGSNDPLFDGSIFQWTQLIPERDINVQLHGETEDLKWSEYLNGSIPL 314
Query: 666 LEAAQNQTP--------SKHLVPDTLGAMLP-HSKQQEEPSQSSSIFSKDLQK 535
Q+Q P + D L L H +QQ++ Q+S+ + KD Q+
Sbjct: 315 STELQSQNPELCNGMKAEEQFGIDGLSIWLQNHQQQQQQQQQASNNYGKDSQR 367
>dbj|BAB39972.1| putative transcription factor (myb) [Oryza sativa (japonica
cultivar-group)] gi|13486753|dbj|BAB39987.1| putative
transcription factor (myb) [Oryza sativa (japonica
cultivar-group)]
Length = 428
Score = 39.7 bits (91), Expect = 0.059
Identities = 33/136 (24%), Positives = 63/136 (46%), Gaps = 21/136 (15%)
Frame = -1
Query: 855 NGSNYWEAS--ASNNSNSSNTSDGIGSRELSSSNILSSSSWGLTDCSSST-------EEA 703
N YW+ + +S++S S+ ++G+G S+S++L ++ + +D + EE
Sbjct: 294 NSQFYWDTNNPSSSSSTGSSGNNGLGFELQSTSSLLETNIFPWSDLAPEKDSQAQLEEEL 353
Query: 702 KWCEYLQNPMLMLEAAQNQTPSKHLVPDTLGAMLPHSKQ------------QEEPSQSSS 559
KW + L + A Q S+ L D + A + + Q+ S
Sbjct: 354 KWPDLLHGTFSEMPAPM-QNLSQSLYEDVVKAESQFNMEGLCAAWSQNLLPQQHLPVVSD 412
Query: 558 IFSKDLQKLTAAFENI 511
++ KDLQ+++ +FENI
Sbjct: 413 MYDKDLQRMSLSFENI 428
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 763,224,268
Number of Sequences: 1393205
Number of extensions: 16965021
Number of successful extensions: 55583
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 43276
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50946
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 45988509105
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)