Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004484A_C01 KMC004484A_c01
(599 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
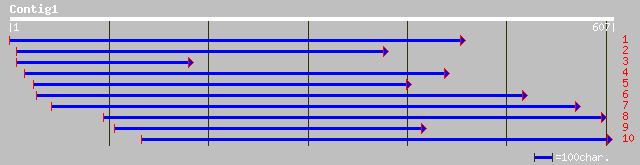
gb|AAL77445.1|L78468_1 acyl-ACP thioesterase [Perilla frutescens] 109 2e-23
gb|AAC72883.1| thioesterase FatA1 [Cuphea hookeriana] 107 1e-22
pir||T07177 probable oleoyl-[acyl-carrier-protein] hydrolase (EC... 106 3e-22
gb|AAB51523.1| acyl-ACP thioesterase [Garcinia mangostana] 104 8e-22
gb|AAL77443.1|L78467_1 acyl-ACP thioesterase [Iris tectorum] 102 4e-21
>gb|AAL77445.1|L78468_1 acyl-ACP thioesterase [Perilla frutescens]
Length = 368
Score = 109 bits (273), Expect = 2e-23
Identities = 58/89 (65%), Positives = 68/89 (76%), Gaps = 2/89 (2%)
Frame = -2
Query: 598 IIDSHELRSITLDYRRECQQSDIVDSLTSVELIEGSEV--VHGTNGSASAAIAQQDKQDR 425
IIDSHEL++ITLDYRRECQ D+VDSLTS E S V + GTNGS +AA +D+ D
Sbjct: 282 IIDSHELQTITLDYRRECQHDDVVDSLTSPESAVNSAVSGLQGTNGSPTAA---KDENDY 338
Query: 424 QQFLHLLRLSTEGLEINRGRTEWRKKAPR 338
QFLHLLR+ST+G EINRGRTEWRKK +
Sbjct: 339 LQFLHLLRISTDGSEINRGRTEWRKKTAK 367
>gb|AAC72883.1| thioesterase FatA1 [Cuphea hookeriana]
Length = 376
Score = 107 bits (267), Expect = 1e-22
Identities = 58/90 (64%), Positives = 69/90 (76%), Gaps = 3/90 (3%)
Frame = -2
Query: 598 IIDSHELRSITLDYRRECQQSDIVDSLTSVELIEGSEV---VHGTNGSASAAIAQQDKQD 428
IIDS+EL +ITLDYRRECQQ D+VDSLTSV E S + GTNGSAS + ++D
Sbjct: 288 IIDSYELETITLDYRRECQQDDVVDSLTSVLSDEESGTLPELKGTNGSASTPL-KRDHDG 346
Query: 427 RQQFLHLLRLSTEGLEINRGRTEWRKKAPR 338
+QFLHLLRLS +GLEINRGRTEWRKK+ +
Sbjct: 347 SRQFLHLLRLSPDGLEINRGRTEWRKKSTK 376
>pir||T07177 probable oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14) -
potato (fragment) gi|2894122|emb|CAA06001.1| acyl
carrier protein thioesterase [Solanum tuberosum]
Length = 118
Score = 106 bits (264), Expect = 3e-22
Identities = 55/89 (61%), Positives = 69/89 (76%), Gaps = 2/89 (2%)
Frame = -2
Query: 598 IIDSHELRSITLDYRRECQQSDIVDSLTSVELIEGSEVV--HGTNGSASAAIAQQDKQDR 425
II++HEL +IT+DYRRECQQ D+VDSLTSVE IE ++ + HG NGSA+A K
Sbjct: 34 IINTHELETITIDYRRECQQDDVVDSLTSVEPIEDADALESHGANGSATA-----PKDVN 88
Query: 424 QQFLHLLRLSTEGLEINRGRTEWRKKAPR 338
+ FLHLLRLS++G+EINR RTEWRKK+ R
Sbjct: 89 KSFLHLLRLSSDGVEINRCRTEWRKKSAR 117
>gb|AAB51523.1| acyl-ACP thioesterase [Garcinia mangostana]
Length = 352
Score = 104 bits (260), Expect = 8e-22
Identities = 53/87 (60%), Positives = 62/87 (70%)
Frame = -2
Query: 598 IIDSHELRSITLDYRRECQQSDIVDSLTSVELIEGSEVVHGTNGSASAAIAQQDKQDRQQ 419
IID+HEL++ITLDYRRECQ D+VDSLTS E E +E V NG+ +A + +
Sbjct: 266 IIDTHELQTITLDYRRECQHDDVVDSLTSPEPSEDAEAVFNHNGTNGSANVSANDHGCRN 325
Query: 418 FLHLLRLSTEGLEINRGRTEWRKKAPR 338
FLHLLRLS GLEINRGRTEWRKK R
Sbjct: 326 FLHLLRLSGNGLEINRGRTEWRKKPTR 352
>gb|AAL77443.1|L78467_1 acyl-ACP thioesterase [Iris tectorum]
Length = 371
Score = 102 bits (254), Expect = 4e-21
Identities = 54/86 (62%), Positives = 69/86 (79%), Gaps = 3/86 (3%)
Frame = -2
Query: 598 IIDSHELRSITLDYRRECQQSDIVDSLTSVELI-EGSEVVH--GTNGSASAAIAQQDKQD 428
IID+HEL++ITLDYRRECQ D+VDSLTS+E++ + + ++H GTNGSA+ ++ QD
Sbjct: 283 IIDTHELQTITLDYRRECQHDDLVDSLTSLEVVGDENGLLHPDGTNGSATG---KRHGQD 339
Query: 427 RQQFLHLLRLSTEGLEINRGRTEWRK 350
+ FLHLLRLS GLEINRGRTEWRK
Sbjct: 340 QCHFLHLLRLSNTGLEINRGRTEWRK 365
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 474,657,026
Number of Sequences: 1393205
Number of extensions: 9555519
Number of successful extensions: 23046
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 22252
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22991
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)