Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004456A_C01 KMC004456A_c01
(568 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
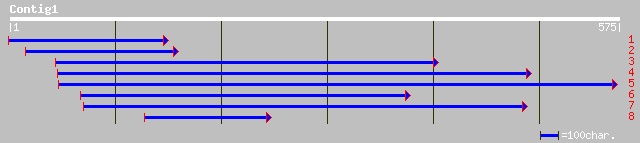
pir||C84599 probable ubiquitin fusion-degradation protein [impor... 133 1e-30
ref|NP_565504.1| putative ubiquitin fusion-degradation protein; ... 133 1e-30
gb|AAM64897.1| putative ubiquitin fusion-degradation protein [Ar... 133 1e-30
ref|NP_568048.1| putative ubiquitin-dependent proteolytic protei... 107 8e-23
pir||T06053 probable ubiquitin-dependent proteolytic protein - A... 105 5e-22
>pir||C84599 probable ubiquitin fusion-degradation protein [imported] -
Arabidopsis thaliana
Length = 320
Score = 133 bits (335), Expect = 1e-30
Identities = 78/127 (61%), Positives = 86/127 (67%), Gaps = 5/127 (3%)
Frame = -2
Query: 567 SAGKAPAAAQETPAETEPKFNPFTGAGRRLDGKPLKYEPPPVSSSGTKDKKPDVPSVNSQ 388
SA K PA A+E E EPKFNPFTG+GRRLDG+PL YEP P SSS K K+P V + N Q
Sbjct: 198 SAAKGPAKAEEVVDEPEPKFNPFTGSGRRLDGRPLAYEPAPASSS--KGKQPVVANGNGQ 255
Query: 387 SSTASSSQSSANQTQGKLVFGSNPAR-----TKDTGKAKEAAKPEPPKEKDEPKFQPFSG 223
SS ASSS+ A + QGKLVFG+N R G AKE K E K KDEPKFQ FSG
Sbjct: 256 SSVASSSE-KATRAQGKLVFGANGNRAPKEAAPKVGAAKETKKEEQEK-KDEPKFQAFSG 313
Query: 222 KKYSLRG 202
KKYSLRG
Sbjct: 314 KKYSLRG 320
>ref|NP_565504.1| putative ubiquitin fusion-degradation protein; protein id:
At2g21270.1, supported by cDNA: 34470. [Arabidopsis
thaliana] gi|20197912|gb|AAD23675.2| putative ubiquitin
fusion-degradation protein [Arabidopsis thaliana]
Length = 319
Score = 133 bits (335), Expect = 1e-30
Identities = 78/127 (61%), Positives = 86/127 (67%), Gaps = 5/127 (3%)
Frame = -2
Query: 567 SAGKAPAAAQETPAETEPKFNPFTGAGRRLDGKPLKYEPPPVSSSGTKDKKPDVPSVNSQ 388
SA K PA A+E E EPKFNPFTG+GRRLDG+PL YEP P SSS K K+P V + N Q
Sbjct: 197 SAAKGPAKAEEVVDEPEPKFNPFTGSGRRLDGRPLAYEPAPASSS--KGKQPVVANGNGQ 254
Query: 387 SSTASSSQSSANQTQGKLVFGSNPAR-----TKDTGKAKEAAKPEPPKEKDEPKFQPFSG 223
SS ASSS+ A + QGKLVFG+N R G AKE K E K KDEPKFQ FSG
Sbjct: 255 SSVASSSE-KATRAQGKLVFGANGNRAPKEAAPKVGAAKETKKEEQEK-KDEPKFQAFSG 312
Query: 222 KKYSLRG 202
KKYSLRG
Sbjct: 313 KKYSLRG 319
>gb|AAM64897.1| putative ubiquitin fusion-degradation protein [Arabidopsis
thaliana]
Length = 319
Score = 133 bits (335), Expect = 1e-30
Identities = 77/127 (60%), Positives = 86/127 (67%), Gaps = 5/127 (3%)
Frame = -2
Query: 567 SAGKAPAAAQETPAETEPKFNPFTGAGRRLDGKPLKYEPPPVSSSGTKDKKPDVPSVNSQ 388
+A K PA A+E E EPKFNPFTG+GRRLDG+PL YEP P SSS KDK+P V + N Q
Sbjct: 197 AAAKGPAKAEEVVDEPEPKFNPFTGSGRRLDGRPLAYEPAPASSS--KDKQPVVANGNGQ 254
Query: 387 SSTASSSQSSANQTQGKLVFGSNPAR-----TKDTGKAKEAAKPEPPKEKDEPKFQPFSG 223
SS ASSS+ A + QGKLVFG+N R G AKE K E K DEPKFQ FSG
Sbjct: 255 SSVASSSE-KATRAQGKLVFGANANRAPKEAAPKVGAAKETKKEEQEK-NDEPKFQAFSG 312
Query: 222 KKYSLRG 202
KKYSLRG
Sbjct: 313 KKYSLRG 319
>ref|NP_568048.1| putative ubiquitin-dependent proteolytic protein; protein id:
At4g38930.1, supported by cDNA: gi_16930496 [Arabidopsis
thaliana] gi|16930497|gb|AAL31934.1|AF419602_1
AT4g38930/F19H22_30 [Arabidopsis thaliana]
gi|22135779|gb|AAM91046.1| AT4g38930/F19H22_30
[Arabidopsis thaliana]
Length = 311
Score = 107 bits (268), Expect = 8e-23
Identities = 66/122 (54%), Positives = 79/122 (64%), Gaps = 4/122 (3%)
Frame = -2
Query: 555 APAAAQETP-AETEPKFNPFTGAGRRLDGKPLKYEPPPVSSSGTKDKKPDVPSVNSQSST 379
APA A+E AE EPKFNPFTG+GRRLDG+PL YEP P +++ P SS+
Sbjct: 197 APAKAKEVDVAEAEPKFNPFTGSGRRLDGRPLSYEPQPGAANSNGQSHP------VASSS 250
Query: 378 ASSSQSSANQTQGKLVFGSNPAR-TKDTGK--AKEAAKPEPPKEKDEPKFQPFSGKKYSL 208
+S S+ + Q +GKLVFGSN R TK+T K A + K E EK E KFQ FSGKKYSL
Sbjct: 251 SSGSEKATQQNRGKLVFGSNVERSTKETTKVGAGKDRKQEEEAEK-EAKFQAFSGKKYSL 309
Query: 207 RG 202
RG
Sbjct: 310 RG 311
>pir||T06053 probable ubiquitin-dependent proteolytic protein - Arabidopsis
thaliana gi|4539312|emb|CAB38813.1| putative
ubiquitin-dependent proteolytic protein [Arabidopsis
thaliana] gi|7270876|emb|CAB80556.1| putative
ubiquitin-dependent proteolytic protein [Arabidopsis
thaliana]
Length = 315
Score = 105 bits (261), Expect = 5e-22
Identities = 65/123 (52%), Positives = 78/123 (62%), Gaps = 4/123 (3%)
Frame = -2
Query: 558 KAPAAAQETP-AETEPKFNPFTGAGRRLDGKPLKYEPPPVSSSGTKDKKPDVPSVNSQSS 382
K A A+E AE EPKFNPFTG+GRRLDG+PL YEP P +++ P SS
Sbjct: 200 KGEAKAKEVDVAEAEPKFNPFTGSGRRLDGRPLSYEPQPGAANSNGQSHP------VASS 253
Query: 381 TASSSQSSANQTQGKLVFGSNPAR-TKDTGK--AKEAAKPEPPKEKDEPKFQPFSGKKYS 211
++S S+ + Q +GKLVFGSN R TK+T K A + K E EK E KFQ FSGKKYS
Sbjct: 254 SSSGSEKATQQNRGKLVFGSNVERSTKETTKVGAGKDRKQEEEAEK-EAKFQAFSGKKYS 312
Query: 210 LRG 202
LRG
Sbjct: 313 LRG 315
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 502,387,327
Number of Sequences: 1393205
Number of extensions: 10839009
Number of successful extensions: 51014
Number of sequences better than 10.0: 526
Number of HSP's better than 10.0 without gapping: 42238
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48316
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)