Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004444A_C02 KMC004444A_c02
(899 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
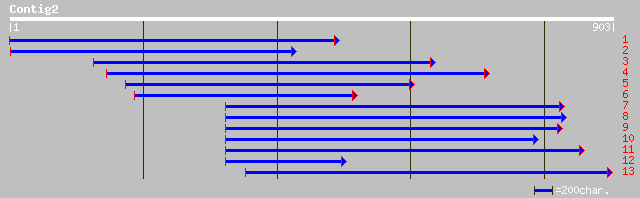
gb|AAM83095.1|AF525402_1 SOS2-like protein kinase [Glycine max] 233 4e-60
gb|AAL37170.1|AF319169_1 CBL-interacting protein kinase [Brassic... 196 3e-49
ref|NP_194825.1| putative protein kinase; protein id: At4g30960.... 195 7e-49
gb|AAD31900.1|AF145482_1 putative serine/threonine protein kinas... 194 2e-48
dbj|BAA92972.1| unnamed protein product [Oryza sativa (japonica ... 154 2e-36
>gb|AAM83095.1|AF525402_1 SOS2-like protein kinase [Glycine max]
Length = 438
Score = 233 bits (593), Expect = 4e-60
Identities = 125/151 (82%), Positives = 136/151 (89%), Gaps = 5/151 (3%)
Frame = -1
Query: 899 EKLKLQEQ--EQVQEQPSTMNAFHIISLSEGFDLSPLFEERKREEKEEMRFATTRPASSV 726
E+L L+E+ + QE +TMNAFHIISLSEGFDLSPLFEE+KREEKE +RFATTRPASSV
Sbjct: 289 EELDLEEKIKQHEQEVSTTMNAFHIISLSEGFDLSPLFEEKKREEKE-LRFATTRPASSV 347
Query: 725 ISRLEEVAKAGKFDVKKSEGKVRLQGQERGRKGKLAIAADMYAVTPSFLVVEVKKDNGDT 546
ISRLE++AKA KFDVKKSE KVRLQGQE+GRKGKLAIAAD+YAVTPSFLVVEVKKDNGDT
Sbjct: 348 ISRLEDLAKAVKFDVKKSETKVRLQGQEKGRKGKLAIAADLYAVTPSFLVVEVKKDNGDT 407
Query: 545 LEYNQFCSKELRPALKDIVWRT---ETPTPA 462
LEYNQFCSKELRPALKDIVWRT E PT A
Sbjct: 408 LEYNQFCSKELRPALKDIVWRTSPAENPTLA 438
>gb|AAL37170.1|AF319169_1 CBL-interacting protein kinase [Brassica napus]
Length = 441
Score = 196 bits (499), Expect = 3e-49
Identities = 96/133 (72%), Positives = 116/133 (87%), Gaps = 1/133 (0%)
Frame = -1
Query: 866 QEQPSTMNAFHIISLSEGFDLSPLFEERKREEKEEMRFATTRPASSVISRLEEVAKAG-K 690
+E+ T+NAFHII+LSEGFDLSPLFEE+K+EEK EMRFAT+RPASSVIS LEE AK G K
Sbjct: 308 KEETETLNAFHIIALSEGFDLSPLFEEKKKEEKMEMRFATSRPASSVISSLEEAAKVGNK 367
Query: 689 FDVKKSEGKVRLQGQERGRKGKLAIAADMYAVTPSFLVVEVKKDNGDTLEYNQFCSKELR 510
FDV+KSE +VR++G++ GRKGKLA+ A+++AV PSF+VVEVKKD+GDTLEYN FCS LR
Sbjct: 368 FDVRKSECRVRIEGKQNGRKGKLAVEAEIFAVAPSFVVVEVKKDHGDTLEYNNFCSTALR 427
Query: 509 PALKDIVWRTETP 471
PALKDI W + TP
Sbjct: 428 PALKDIFWTSTTP 440
>ref|NP_194825.1| putative protein kinase; protein id: At4g30960.1, supported by
cDNA: gi_13448032, supported by cDNA: gi_14334745,
supported by cDNA: gi_15293234, supported by cDNA:
gi_16930694, supported by cDNA: gi_9280633 [Arabidopsis
thaliana] gi|25387050|pir||E85362 hypothetical protein
AT4g30960 [imported] - Arabidopsis thaliana
gi|2980770|emb|CAA18197.1| putative protein kinase
[Arabidopsis thaliana] gi|7269998|emb|CAB79814.1|
putative protein kinase [Arabidopsis thaliana]
gi|9280634|gb|AAF86505.1|AF285106_1 CBL-interacting
protein kinase 6 [Arabidopsis thaliana]
gi|13448033|gb|AAK26843.1|AF339145_1 SOS2-like protein
kinase PKS4 [Arabidopsis thaliana]
gi|14334746|gb|AAK59551.1| putative protein kinase
[Arabidopsis thaliana] gi|15293235|gb|AAK93728.1|
putative protein kinase [Arabidopsis thaliana]
gi|16930695|gb|AAL32013.1|AF436831_1 AT4g30960/F6I18_130
[Arabidopsis thaliana]
Length = 441
Score = 195 bits (496), Expect = 7e-49
Identities = 97/136 (71%), Positives = 117/136 (85%), Gaps = 1/136 (0%)
Frame = -1
Query: 866 QEQPSTMNAFHIISLSEGFDLSPLFEERKREEKEEMRFATTRPASSVISRLEEVAKAG-K 690
+E+ T+NAFHII+LSEGFDLSPLFEE+K+EEK EMRFAT+RPASSVIS LEE A+ G K
Sbjct: 309 KEETETLNAFHIIALSEGFDLSPLFEEKKKEEKREMRFATSRPASSVISSLEEAARVGNK 368
Query: 689 FDVKKSEGKVRLQGQERGRKGKLAIAADMYAVTPSFLVVEVKKDNGDTLEYNQFCSKELR 510
FDV+KSE +VR++G++ GRKGKLA+ A+++AV PSF+VVEVKKD+GDTLEYN FCS LR
Sbjct: 369 FDVRKSESRVRIEGKQNGRKGKLAVEAEIFAVAPSFVVVEVKKDHGDTLEYNNFCSTALR 428
Query: 509 PALKDIVWRTETPTPA 462
PALKDI W T TPA
Sbjct: 429 PALKDIFW---TSTPA 441
>gb|AAD31900.1|AF145482_1 putative serine/threonine protein kinase [Mesembryanthemum
crystallinum]
Length = 462
Score = 194 bits (492), Expect = 2e-48
Identities = 98/130 (75%), Positives = 117/130 (89%), Gaps = 3/130 (2%)
Frame = -1
Query: 866 QEQPSTMNAFHIISLSEGFDLSPLFEERKREEKEEMRFATTRPASSVISRLEEVA-KAGK 690
+++P +NAFHIISLS+GFDLSPLFEE+ REE+++MRFATT+ AS+V+SRLEEVA KAGK
Sbjct: 328 KKEPEMLNAFHIISLSQGFDLSPLFEEKHREERQDMRFATTKSASTVVSRLEEVAAKAGK 387
Query: 689 FDVKK--SEGKVRLQGQERGRKGKLAIAADMYAVTPSFLVVEVKKDNGDTLEYNQFCSKE 516
F VKK + VRLQGQERGRKGKLAIAA++ +V+PS LVVEVKKDNGDTLEYNQFC+K+
Sbjct: 388 FIVKKFCGDSVVRLQGQERGRKGKLAIAAEIMSVSPSLLVVEVKKDNGDTLEYNQFCTKQ 447
Query: 515 LRPALKDIVW 486
LRPALKDIVW
Sbjct: 448 LRPALKDIVW 457
>dbj|BAA92972.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|20804455|dbj|BAB92151.1|
ATF6I18_13(AL022198|pid:g2980770) Arabidopsis thaliana
chromosome 4 BAC clone F6I18 ; putative protein kinase.
[Oryza sativa (japonica cultivar-group)]
Length = 461
Score = 154 bits (388), Expect = 2e-36
Identities = 75/133 (56%), Positives = 103/133 (77%)
Frame = -1
Query: 860 QPSTMNAFHIISLSEGFDLSPLFEERKREEKEEMRFATTRPASSVISRLEEVAKAGKFDV 681
+P ++NAF IISLS+GFDLS LFE K E+K + RF T +PAS+++S+LE++A+ F V
Sbjct: 311 KPVSLNAFDIISLSKGFDLSGLFENDK-EQKADSRFMTQKPASAIVSKLEQIAETESFKV 369
Query: 680 KKSEGKVRLQGQERGRKGKLAIAADMYAVTPSFLVVEVKKDNGDTLEYNQFCSKELRPAL 501
KK +G V+LQG + GRKG+LAI A+++ VTPSF VVEVKK GDTLEY +FC+K LRP+L
Sbjct: 370 KKQDGLVKLQGSKEGRKGQLAIDAEIFEVTPSFFVVEVKKSAGDTLEYEKFCNKGLRPSL 429
Query: 500 KDIVWRTETPTPA 462
+DI W ++ P+
Sbjct: 430 RDICWDGQSEHPS 442
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 804,530,229
Number of Sequences: 1393205
Number of extensions: 20339326
Number of successful extensions: 167717
Number of sequences better than 10.0: 3530
Number of HSP's better than 10.0 without gapping: 89132
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 133577
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 49333127949
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)