Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004439A_C01 KMC004439A_c01
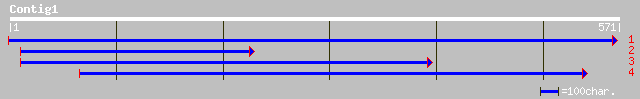
(560 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM65587.1| unknown [Arabidopsis thaliana] 137 7e-32
ref|NP_565365.1| Expressed protein; protein id: At2g14835.1, sup... 137 7e-32
gb|AAK74006.1| At2g14841 [Arabidopsis thaliana] gi|16323316|gb|A... 135 4e-31
gb|AAK92653.1|AC079634_14 Hypothetical protein [Oryza sativa] 84 2e-15
sp|Q96624|VCO7_ADEB2 Major core protein precursor (Protein VII) ... 33 3.1
>gb|AAM65587.1| unknown [Arabidopsis thaliana]
Length = 343
Score = 137 bits (346), Expect = 7e-32
Identities = 66/84 (78%), Positives = 75/84 (88%)
Frame = -2
Query: 559 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACM 380
FLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQRSS+MD RKIL+ IA++ACM
Sbjct: 259 FLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQRSSKMDIRKILIFIALIACM 318
Query: 379 ATMGILYYRLVQRGPGEELPNEEQ 308
ATMGILYYRL + G+ELP+EEQ
Sbjct: 319 ATMGILYYRLALQAIGQELPDEEQ 342
>ref|NP_565365.1| Expressed protein; protein id: At2g14835.1, supported by cDNA:
40603., supported by cDNA: gi_15010693, supported by
cDNA: gi_15983469, supported by cDNA: gi_16323315
[Arabidopsis thaliana]
gi|15983470|gb|AAL11603.1|AF424609_1 unknown protein
[Arabidopsis thaliana] gi|20197262|gb|AAM15000.1|
Expressed protein [Arabidopsis thaliana]
Length = 343
Score = 137 bits (346), Expect = 7e-32
Identities = 66/84 (78%), Positives = 75/84 (88%)
Frame = -2
Query: 559 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACM 380
FLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQRSS+MD RKIL+ IA++ACM
Sbjct: 259 FLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQRSSKMDIRKILIFIALIACM 318
Query: 379 ATMGILYYRLVQRGPGEELPNEEQ 308
ATMGILYYRL + G+ELP+EEQ
Sbjct: 319 ATMGILYYRLALQAIGQELPDEEQ 342
>gb|AAK74006.1| At2g14841 [Arabidopsis thaliana] gi|16323316|gb|AAL15413.1|
At2g14841/At2g14841 [Arabidopsis thaliana]
Length = 343
Score = 135 bits (339), Expect = 4e-31
Identities = 65/84 (77%), Positives = 74/84 (87%)
Frame = -2
Query: 559 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACM 380
FLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQRSS+MD KIL+ IA++ACM
Sbjct: 259 FLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQRSSKMDIGKILIFIALIACM 318
Query: 379 ATMGILYYRLVQRGPGEELPNEEQ 308
ATMGILYYRL + G+ELP+EEQ
Sbjct: 319 ATMGILYYRLALQAIGQELPDEEQ 342
>gb|AAK92653.1|AC079634_14 Hypothetical protein [Oryza sativa]
Length = 383
Score = 83.6 bits (205), Expect = 2e-15
Identities = 41/57 (71%), Positives = 48/57 (83%)
Frame = -2
Query: 559 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIM 389
FLR LLPFWSSALPTLPVTAPPRK+ ++ EGR+RHQ+SSRMDP KILL +AI+
Sbjct: 295 FLRMLLPFWSSALPTLPVTAPPRKE----SDAPEGRSRHQKSSRMDPTKILLALAII 347
>sp|Q96624|VCO7_ADEB2 Major core protein precursor (Protein VII) (pVII)
gi|1574960|gb|AAB16756.1| major core protein precursor
[bovine adenovirus 2]
Length = 183
Score = 32.7 bits (73), Expect = 3.1
Identities = 21/64 (32%), Positives = 32/64 (49%)
Frame = -2
Query: 529 SALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACMATMGILYYRL 350
+ +PT+P+T P D NA +EG TR +R + R+ A+ A A + RL
Sbjct: 50 TTVPTVPITDDPVADVVNAI--AEGTTRRRRRAERRRRRRQATSAMRAARALVRSARRRL 107
Query: 349 VQRG 338
+RG
Sbjct: 108 ARRG 111
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 477,275,713
Number of Sequences: 1393205
Number of extensions: 10024470
Number of successful extensions: 24007
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 23353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23999
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)