Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004427A_C01 KMC004427A_c01
(943 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
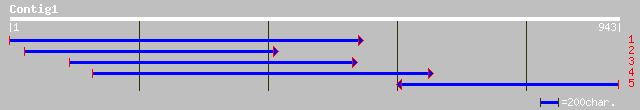
ref|NP_564249.1| 14-3-3 protein GF14 iota (grf12); protein id: A... 276 3e-73
pir||F86391 T1K7.15 protein - Arabidopsis thaliana gi|9797752|gb... 276 3e-73
gb|AAM62569.1| 14-3-3-like protein GF14 iota (General regulatory... 276 3e-73
gb|AAL28067.1|AF426249_1 14-3-3 protein [Fritillaria cirrhosa] 271 8e-72
gb|AAD27824.2| 14-3-3 protein [Populus x canescens] 244 1e-63
>ref|NP_564249.1| 14-3-3 protein GF14 iota (grf12); protein id: At1g26480.1,
supported by cDNA: 146580., supported by cDNA:
gi_12963452 [Arabidopsis thaliana]
gi|20137460|sp|Q9C5W6|143C_ARATH 14-3-3-like protein
GF14 iota (General regulatory factor 12)
gi|12963453|gb|AAK11271.1|AF335544_1 14-3-3 protein
GF14iota [Arabidopsis thaliana]
gi|26450876|dbj|BAC42545.1| putative 14-3-3 protein
epsilon [Arabidopsis thaliana]
Length = 268
Score = 276 bits (707), Expect = 3e-73
Identities = 138/177 (77%), Positives = 152/177 (84%)
Frame = -1
Query: 943 GNEHKLSRSRITAKRLRRNSPKFVVTSWTIIDQHLIPSSASAEASVFYYKMKGDYYRYLA 764
GNE + + + +++ TIIDQHLIP + S EA+VFYYKMKGDYYRYLA
Sbjct: 80 GNESNVKQIKGYRQKVEDELANICQDILTIIDQHLIPHATSGEATVFYYKMKGDYYRYLA 139
Query: 763 EFKTDQERKEAAEQSLKGYEAASATANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH 584
EFKT+QERKEAAEQSLKGYEAA+ A+T+LPSTHPIRLGLALNFSVFYYEIMNSPERACH
Sbjct: 140 EFKTEQERKEAAEQSLKGYEAATQAASTELPSTHPIRLGLALNFSVFYYEIMNSPERACH 199
Query: 583 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEENIKAEEAK 413
LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGE+NIK EE+K
Sbjct: 200 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEDNIKTEESK 256
>pir||F86391 T1K7.15 protein - Arabidopsis thaliana
gi|9797752|gb|AAF98570.1|AC013427_13 Strong similarity
to GF14 mu from Arabidopsis thaliana gb|AB011545 and is
a member of the 14-3-3 protein PF|00244 family
Length = 286
Score = 276 bits (707), Expect = 3e-73
Identities = 138/177 (77%), Positives = 152/177 (84%)
Frame = -1
Query: 943 GNEHKLSRSRITAKRLRRNSPKFVVTSWTIIDQHLIPSSASAEASVFYYKMKGDYYRYLA 764
GNE + + + +++ TIIDQHLIP + S EA+VFYYKMKGDYYRYLA
Sbjct: 80 GNESNVKQIKGYRQKVEDELANICQDILTIIDQHLIPHATSGEATVFYYKMKGDYYRYLA 139
Query: 763 EFKTDQERKEAAEQSLKGYEAASATANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH 584
EFKT+QERKEAAEQSLKGYEAA+ A+T+LPSTHPIRLGLALNFSVFYYEIMNSPERACH
Sbjct: 140 EFKTEQERKEAAEQSLKGYEAATQAASTELPSTHPIRLGLALNFSVFYYEIMNSPERACH 199
Query: 583 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEENIKAEEAK 413
LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGE+NIK EE+K
Sbjct: 200 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEDNIKTEESK 256
>gb|AAM62569.1| 14-3-3-like protein GF14 iota (General regulatory factor 12)
[Arabidopsis thaliana]
Length = 253
Score = 276 bits (707), Expect = 3e-73
Identities = 138/177 (77%), Positives = 152/177 (84%)
Frame = -1
Query: 943 GNEHKLSRSRITAKRLRRNSPKFVVTSWTIIDQHLIPSSASAEASVFYYKMKGDYYRYLA 764
GNE + + + +++ TIIDQHLIP + S EA+VFYYKMKGDYYRYLA
Sbjct: 65 GNESNVKQIKGYRQKVEDELANICQDILTIIDQHLIPHATSGEATVFYYKMKGDYYRYLA 124
Query: 763 EFKTDQERKEAAEQSLKGYEAASATANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH 584
EFKT+QERKEAAEQSLKGYEAA+ A+T+LPSTHPIRLGLALNFSVFYYEIMNSPERACH
Sbjct: 125 EFKTEQERKEAAEQSLKGYEAATQAASTELPSTHPIRLGLALNFSVFYYEIMNSPERACH 184
Query: 583 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEENIKAEEAK 413
LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGE+NIK EE+K
Sbjct: 185 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEDNIKTEESK 241
>gb|AAL28067.1|AF426249_1 14-3-3 protein [Fritillaria cirrhosa]
Length = 262
Score = 271 bits (694), Expect = 8e-72
Identities = 135/177 (76%), Positives = 148/177 (83%)
Frame = -1
Query: 943 GNEHKLSRSRITAKRLRRNSPKFVVTSWTIIDQHLIPSSASAEASVFYYKMKGDYYRYLA 764
GN+ + + ++ K T+ID+HLIPSS S EA+VFYYKMKGDYYRYLA
Sbjct: 79 GNDQNVKLIKGYRHKVEEELAKICNDILTVIDKHLIPSSGSGEATVFYYKMKGDYYRYLA 138
Query: 763 EFKTDQERKEAAEQSLKGYEAASATANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH 584
EFK + ERKEAA+QS+K Y+AAS TANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH
Sbjct: 139 EFKNEHERKEAADQSMKAYQAASNTANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH 198
Query: 583 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEENIKAEEAK 413
LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGG+E K EEAK
Sbjct: 199 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGDEGFKGEEAK 255
>gb|AAD27824.2| 14-3-3 protein [Populus x canescens]
Length = 260
Score = 244 bits (624), Expect = 1e-63
Identities = 121/172 (70%), Positives = 140/172 (81%)
Frame = -1
Query: 943 GNEHKLSRSRITAKRLRRNSPKFVVTSWTIIDQHLIPSSASAEASVFYYKMKGDYYRYLA 764
GNE + R + K++ T+ID+HLIPSS E++VFYYKMKGDYYRYLA
Sbjct: 77 GNETNVKRIKEYRKKVEAELTGVCNDIMTVIDEHLIPSSIPGESAVFYYKMKGDYYRYLA 136
Query: 763 EFKTDQERKEAAEQSLKGYEAASATANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACH 584
EFK+ ERKEAA+QSLK YE A++TA DL THPIRLGLALNFSVFYYEIMNSPERACH
Sbjct: 137 EFKSGNERKEAADQSLKAYETATSTAARDLSPTHPIRLGLALNFSVFYYEIMNSPERACH 196
Query: 583 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDLPEDGGEENIK 428
LAKQ+FDEAI+ELDTLSEESYKDSTLIMQLLRDNLTLWTSD+PEDG ++ ++
Sbjct: 197 LAKQSFDEAISELDTLSEESYKDSTLIMQLLRDNLTLWTSDIPEDGEDQKME 248
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 787,009,326
Number of Sequences: 1393205
Number of extensions: 16704830
Number of successful extensions: 47492
Number of sequences better than 10.0: 327
Number of HSP's better than 10.0 without gapping: 45166
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 47288
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 52691756080
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)