Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004381A_C01 KMC004381A_c01
(594 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
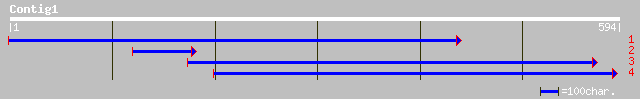
ref|NP_563990.1| ovule development protein, putative; protein id... 51 1e-05
pir||D86295 hypothetical protein T24D18.16 - Arabidopsis thalian... 51 1e-05
ref|NP_178088.1| ovule development protein, putative; protein id... 40 0.022
ref|NP_702855.1| erythrocyte membrane protein 1 (PfEMP1) [Plasmo... 32 4.7
ref|NP_629174.1| putative lipoprotein [Streptomyces coelicolor A... 32 6.1
>ref|NP_563990.1| ovule development protein, putative; protein id: At1g16060.1,
supported by cDNA: gi_15028184 [Arabidopsis thaliana]
gi|15028185|gb|AAK76589.1| unknown protein [Arabidopsis
thaliana] gi|22136940|gb|AAM91814.1| unknown protein
[Arabidopsis thaliana]
Length = 345
Score = 51.2 bits (121), Expect = 1e-05
Identities = 26/46 (56%), Positives = 32/46 (69%), Gaps = 1/46 (2%)
Frame = -3
Query: 475 PRRTFPEDIQTIFENEDSSIYT-ENDDIIFGDLSSIAAPIFHFELD 341
PRR+FPEDIQT F ++S T E DD+IFGDL S P F+ EL+
Sbjct: 298 PRRSFPEDIQTYFGCQNSGKLTAEEDDVIFGDLDSFLTPDFYSELN 343
>pir||D86295 hypothetical protein T24D18.16 - Arabidopsis thaliana
gi|6587812|gb|AAF18503.1|AC010924_16 Similar to
gb|U44028 transcription factor CKC from Arabidopsis
thaliana and contains two PF|00847 AP2 domains
Length = 332
Score = 51.2 bits (121), Expect = 1e-05
Identities = 26/46 (56%), Positives = 32/46 (69%), Gaps = 1/46 (2%)
Frame = -3
Query: 475 PRRTFPEDIQTIFENEDSSIYT-ENDDIIFGDLSSIAAPIFHFELD 341
PRR+FPEDIQT F ++S T E DD+IFGDL S P F+ EL+
Sbjct: 285 PRRSFPEDIQTYFGCQNSGKLTAEEDDVIFGDLDSFLTPDFYSELN 330
>ref|NP_178088.1| ovule development protein, putative; protein id: At1g79700.1
[Arabidopsis thaliana] gi|25405039|pir||H96827 protein
F20B17.12 [imported] - Arabidopsis thaliana
gi|7715603|gb|AAF68121.1|AC010793_16 F20B17.12
[Arabidopsis thaliana]
Length = 308
Score = 40.0 bits (92), Expect = 0.022
Identities = 22/56 (39%), Positives = 31/56 (55%), Gaps = 5/56 (8%)
Frame = -3
Query: 493 VESTTLP-----RRTFPEDIQTIFENEDSSIYTENDDIIFGDLSSIAAPIFHFELD 341
+E +T P RR+FP+DIQT F +DS +D+IF +S P F+ E D
Sbjct: 250 IEPSTSPEVIPTRRSFPDDIQTYFGCQDSGKLATEEDVIFDCFNSYINPGFYNEFD 305
>ref|NP_702855.1| erythrocyte membrane protein 1 (PfEMP1) [Plasmodium falciparum 3D7]
gi|23498272|emb|CAD49243.1| erythrocyte membrane protein
1 (PfEMP1) [Plasmodium falciparum 3D7]
Length = 2152
Score = 32.3 bits (72), Expect = 4.7
Identities = 16/65 (24%), Positives = 32/65 (48%)
Frame = -3
Query: 451 IQTIFENEDSSIYTENDDIIFGDLSSIAAPIFHFELDT*KRELPAYIYEQCFINYVNFQN 272
I + +N + + DDII+ D+ P++H +D K +P + Q +N +N Q
Sbjct: 2081 IDNLEKNSEPYYDIDEDDIIYFDIDDEKTPMYHNNMDNNKSNVPTKV--QIEMNVINKQE 2138
Query: 271 VINQK 257
+ ++
Sbjct: 2139 LFQEE 2143
>ref|NP_629174.1| putative lipoprotein [Streptomyces coelicolor A3(2)]
gi|8218184|emb|CAB92615.1| putative lipoprotein
[Streptomyces coelicolor A3(2)]
Length = 220
Score = 32.0 bits (71), Expect = 6.1
Identities = 22/64 (34%), Positives = 34/64 (52%), Gaps = 4/64 (6%)
Frame = -3
Query: 517 KTSSALAPVESTTL--PRRTFPEDIQTIFENEDSSIYTENDDIIFGDLS--SIAAPIFHF 350
+TS+A +P E+ T P+ TFP D + +FE E + T++ + LS S+ IF
Sbjct: 47 ETSAAPSPSETDTAGRPKITFPSDAKNVFEYEKTGNATKDAALADSTLSVNSVDEAIFEG 106
Query: 349 ELDT 338
DT
Sbjct: 107 STDT 110
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 424,397,064
Number of Sequences: 1393205
Number of extensions: 8275824
Number of successful extensions: 17992
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 17564
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17983
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22854740960
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)