Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
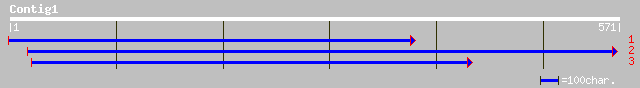
Query= KMC004361A_C01 KMC004361A_c01
(571 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|EAA40145.1| GLP_393_1379_2998 [Giardia lamblia ATCC 50803] 33 3.3
ref|NP_190417.1| putative protein; protein id: At3g48480.1 [Arab... 33 3.3
ref|NP_595975.1| conserved hypothetical protein [Schizosaccharom... 32 4.3
gb|EAA16312.1| asf1 gene product [Plasmodium yoelii yoelii] 32 5.6
ref|NP_567478.1| Expressed protein; protein id: At4g15880.1, sup... 32 7.3
>gb|EAA40145.1| GLP_393_1379_2998 [Giardia lamblia ATCC 50803]
Length = 539
Score = 32.7 bits (73), Expect = 3.3
Identities = 15/34 (44%), Positives = 19/34 (55%)
Frame = -1
Query: 412 NIKCPRQTNNIDCGYYILKFMKDIITHNEPMIPL 311
++ P+Q N DCG Y+L FMK I P PL
Sbjct: 474 SLNMPQQENGYDCGVYMLLFMKMIARQFGPGYPL 507
>ref|NP_190417.1| putative protein; protein id: At3g48480.1 [Arabidopsis thaliana]
gi|7630035|emb|CAB88329.1| putative protein [Arabidopsis
thaliana]
Length = 169
Score = 32.7 bits (73), Expect = 3.3
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Frame = -1
Query: 400 PRQTNNIDCGYYILKFMKDIITHNEPMIPLEYFQDCRCSFYSNDQLDEVREEWASYV-LD 224
P+QTN+++CG ++L ++ + +D +F D ++E+W S+ L+
Sbjct: 109 PQQTNDVECGSFVLYYIH------------RFIEDAPENFNVEDMPYFLKEDWFSHKDLE 156
Query: 223 NFC*PLGSL 197
FC L SL
Sbjct: 157 KFCDELHSL 165
>ref|NP_595975.1| conserved hypothetical protein [Schizosaccharomyces pombe]
gi|15214321|sp|O42957|ULP1_SCHPO Ubiquitin-like-specific
protease 1 gi|7490324|pir||T39840 conserved hypothetical
protein SPBC19G7.09 - fission yeast
(Schizosaccharomyces pombe) gi|2894265|emb|CAA17063.1|
conserved hypothetical protein [Schizosaccharomyces
pombe]
Length = 568
Score = 32.3 bits (72), Expect = 4.3
Identities = 29/116 (25%), Positives = 47/116 (40%)
Frame = -1
Query: 571 NASEGKIYYLDPLHGDPRHXPEMKTLFDTALKIYRVTTSATVPGARRSHIQWDNIKCPRQ 392
N S+ + Y D L G P +FD Y T V + + DN PRQ
Sbjct: 470 NKSKKRFEYWDSLAGSPGK------VFDLLRDYYIAETKGAVDVSDWENFMDDN--SPRQ 521
Query: 391 TNNIDCGYYILKFMKDIITHNEPMIPLEYFQDCRCSFYSNDQLDEVREEWASYVLD 224
N DCG + K + ++ N P+ +S + + E+R + A+ ++D
Sbjct: 522 RNGHDCGVFACK-TAECVSRNVPV------------QFSQNDMPELRIKMAASIID 564
>gb|EAA16312.1| asf1 gene product [Plasmodium yoelii yoelii]
Length = 178
Score = 32.0 bits (71), Expect = 5.6
Identities = 17/55 (30%), Positives = 25/55 (44%)
Frame = -1
Query: 382 IDCGYYILKFMKDIITHNEPMIPLEYFQDCRCSFYSNDQLDEVREEWASYVLDNF 218
I YY+ F KDI +P + +Y + CR F N ++ + W S D F
Sbjct: 24 IRIAYYMNSFYKDIELREKPPVSPQYDKICRHIFVDNPRIVKFSIPWDSEERDEF 78
>ref|NP_567478.1| Expressed protein; protein id: At4g15880.1, supported by cDNA:
gi_14423393, supported by cDNA: gi_20148438 [Arabidopsis
thaliana] gi|14423394|gb|AAK62379.1|AF386934_1 Unknown
protein [Arabidopsis thaliana]
gi|20148439|gb|AAM10110.1| unknown protein [Arabidopsis
thaliana]
Length = 489
Score = 31.6 bits (70), Expect = 7.3
Identities = 24/78 (30%), Positives = 34/78 (42%), Gaps = 4/78 (5%)
Frame = -1
Query: 571 NASEGKIYYLDPLHG-DPRHXPEMKTLFDTALKIYRVTTSATVPGARRSHIQWDNI---K 404
N E K+ YLD L+G DP + AL Y + G + WD
Sbjct: 387 NNRESKLLYLDSLNGVDP--------MILNALAKYMGDEANEKSGKKIDANSWDMEFVED 438
Query: 403 CPRQTNNIDCGYYILKFM 350
P+Q N DCG ++LK++
Sbjct: 439 LPQQKNGYDCGMFMLKYI 456
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 448,594,885
Number of Sequences: 1393205
Number of extensions: 8873195
Number of successful extensions: 22681
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 21979
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22659
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)