Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004334A_C01 KMC004334A_c01
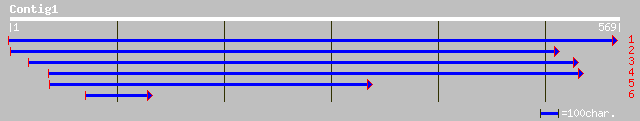
(568 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM64738.1| unknown [Arabidopsis thaliana] 167 1e-40
ref|NP_565538.1| Expressed protein; protein id: At2g22475.1, sup... 167 1e-40
dbj|BAC43153.1| unknown protein [Arabidopsis thaliana] 163 2e-39
ref|NP_174141.1| FH protein interacting protein FIP1; protein id... 140 1e-32
dbj|BAC10185.1| contains EST C91668(E31216)~similar to FH protei... 138 4e-32
>gb|AAM64738.1| unknown [Arabidopsis thaliana]
Length = 299
Score = 167 bits (422), Expect = 1e-40
Identities = 77/100 (77%), Positives = 91/100 (91%)
Frame = -3
Query: 566 LSTSAGPVMGVLYVSTAKIAYSSDNPISYKSENKTEWSYYKVVIPLHELKAVNPSANTAN 387
LSTSAGPVMGVLY+S+AK+AY SDNP+SYK+ ++TEWSYYKVVIPLH+LKAVNPSA+ N
Sbjct: 197 LSTSAGPVMGVLYISSAKLAYCSDNPLSYKNGDQTEWSYYKVVIPLHQLKAVNPSASIVN 256
Query: 386 PAEKYIQVISTDNHEFWFMGFLNYEGAQECLEEALKAGKL 267
PAEKYIQVIS DNHEFWFMGFLNY+GA L+++L+AG L
Sbjct: 257 PAEKYIQVISVDNHEFWFMGFLNYDGAVTSLQDSLQAGAL 296
>ref|NP_565538.1| Expressed protein; protein id: At2g22475.1, supported by cDNA:
32573. [Arabidopsis thaliana] gi|20197888|gb|AAM15301.1|
Expressed protein [Arabidopsis thaliana]
Length = 299
Score = 167 bits (422), Expect = 1e-40
Identities = 77/100 (77%), Positives = 91/100 (91%)
Frame = -3
Query: 566 LSTSAGPVMGVLYVSTAKIAYSSDNPISYKSENKTEWSYYKVVIPLHELKAVNPSANTAN 387
LSTSAGPVMGVLY+S+AK+AY SDNP+SYK+ ++TEWSYYKVVIPLH+LKAVNPSA+ N
Sbjct: 197 LSTSAGPVMGVLYISSAKLAYCSDNPLSYKNGDQTEWSYYKVVIPLHQLKAVNPSASIVN 256
Query: 386 PAEKYIQVISTDNHEFWFMGFLNYEGAQECLEEALKAGKL 267
PAEKYIQVIS DNHEFWFMGFLNY+GA L+++L+AG L
Sbjct: 257 PAEKYIQVISVDNHEFWFMGFLNYDGAVTSLQDSLQAGAL 296
>dbj|BAC43153.1| unknown protein [Arabidopsis thaliana]
Length = 299
Score = 163 bits (412), Expect = 2e-39
Identities = 76/100 (76%), Positives = 90/100 (90%)
Frame = -3
Query: 566 LSTSAGPVMGVLYVSTAKIAYSSDNPISYKSENKTEWSYYKVVIPLHELKAVNPSANTAN 387
LSTSAG VMGVLY+S+AK+AY SDNP+SYK+ ++TEWSYYKVVIPLH+LKAVNPSA+ N
Sbjct: 197 LSTSAGLVMGVLYISSAKLAYCSDNPLSYKNGDQTEWSYYKVVIPLHQLKAVNPSASIVN 256
Query: 386 PAEKYIQVISTDNHEFWFMGFLNYEGAQECLEEALKAGKL 267
PAEKYIQVIS DNHEFWFMGFLNY+GA L+++L+AG L
Sbjct: 257 PAEKYIQVISVDNHEFWFMGFLNYDGAVTSLQDSLQAGAL 296
>ref|NP_174141.1| FH protein interacting protein FIP1; protein id: At1g28200.1,
supported by cDNA: 30245., supported by cDNA:
gi_13877828, supported by cDNA: gi_15912222, supported
by cDNA: gi_16323497, supported by cDNA: gi_6503011
[Arabidopsis thaliana] gi|25344199|pir||A86408 FH
protein interacting protein FIP1 [imported] -
Arabidopsis thaliana gi|6503012|gb|AAF14549.1|AF174428_1
FH protein interacting protein FIP1 [Arabidopsis
thaliana] gi|9795617|gb|AAF98435.1|AC021044_14 FH
protein interacting protein FIP1 [Arabidopsis thaliana]
gi|13877829|gb|AAK43992.1|AF370177_1 putative FH protein
interacting protein FIP1 [Arabidopsis thaliana]
gi|15912223|gb|AAL08245.1| At1g28200/F3H9_12
[Arabidopsis thaliana] gi|16323498|gb|AAL15243.1|
putative FH protein interacting protein FIP1
[Arabidopsis thaliana] gi|21592606|gb|AAM64555.1| FH
protein interacting protein FIP1 [Arabidopsis thaliana]
Length = 259
Score = 140 bits (352), Expect = 1e-32
Identities = 63/97 (64%), Positives = 82/97 (83%)
Frame = -3
Query: 566 LSTSAGPVMGVLYVSTAKIAYSSDNPISYKSENKTEWSYYKVVIPLHELKAVNPSANTAN 387
LSTSAGPV+GV+Y+ST K+A+SSDNP+SYK +T WSYYKVV+P ++LKAVNPS + N
Sbjct: 160 LSTSAGPVLGVMYLSTHKLAFSSDNPLSYKEGEQTLWSYYKVVLPANQLKAVNPSTSRVN 219
Query: 386 PAEKYIQVISTDNHEFWFMGFLNYEGAQECLEEALKA 276
++KYIQVIS DNHEFWFMGF+ YE A + L+EA+++
Sbjct: 220 TSDKYIQVISIDNHEFWFMGFVTYESAVKSLQEAVQS 256
>dbj|BAC10185.1| contains EST C91668(E31216)~similar to FH protein interacting
protein FIP1 [Oryza sativa (japonica cultivar-group)]
Length = 297
Score = 138 bits (348), Expect = 4e-32
Identities = 63/103 (61%), Positives = 87/103 (84%), Gaps = 1/103 (0%)
Frame = -3
Query: 566 LSTSAGPVMGVLYVSTAKIAYSSDNPISYKSE-NKTEWSYYKVVIPLHELKAVNPSANTA 390
LSTS GP+MGVLY+STAKIA+ SD+P++Y +E NK + S YKVV+P+ +L++V P+A+
Sbjct: 188 LSTSHGPIMGVLYISTAKIAFCSDSPVAYVTEDNKNQSSIYKVVVPVAQLRSVTPTASQQ 247
Query: 389 NPAEKYIQVISTDNHEFWFMGFLNYEGAQECLEEALKAGKLSL 261
NPAE+YIQV+S DNH+FWFMGF+NY+GA + L+EA++ GK SL
Sbjct: 248 NPAERYIQVVSVDNHDFWFMGFVNYDGAVKSLQEAVRGGKASL 290
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,119,551
Number of Sequences: 1393205
Number of extensions: 9724655
Number of successful extensions: 19462
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 19112
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19450
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)