Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
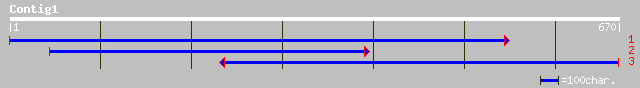
Query= KMC004308A_C01 KMC004308A_c01
(670 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_563751.1| expressed protein; protein id: At1g05840.1, sup... 195 5e-49
ref|NP_198475.1| aspartyl protease-like; protein id: At5g36260.1... 167 1e-40
gb|AAM20468.1| putative aspartyl protease [Arabidopsis thaliana]... 166 2e-40
ref|NP_186923.1| putative aspartyl protease; protein id: At3g027... 166 2e-40
ref|NP_176703.1| hypothetical protein; protein id: At1g65240.1 [... 155 4e-37
>ref|NP_563751.1| expressed protein; protein id: At1g05840.1, supported by cDNA:
158528. [Arabidopsis thaliana]
Length = 485
Score = 195 bits (495), Expect = 5e-49
Identities = 91/155 (58%), Positives = 118/155 (75%)
Frame = -3
Query: 626 IVYDQLMSKVLAKQPRLKVYLVEEQYSCFQYTGNVDSGFPIVKLHFEDSLSLTVYPHDYL 447
I+Y+ L+ K+ +++P LKV++V++ Y CFQY+G VD GFP V HFE+S+ L VYPHDYL
Sbjct: 326 IIYEPLVKKITSQEPALKVHIVDKDYKCFQYSGRVDEGFPNVTFHFENSVFLRVYPHDYL 385
Query: 446 FNYKGDSYWCIGWQKSASETKNGKDMTLLGDFVLSNKLVVYDLENMTIGWTDYNCSSSIK 267
F ++G WCIGWQ SA ++++ ++MTLLGD VLSNKLV+YDLEN IGWT+YNCSSSIK
Sbjct: 386 FPHEG--MWCIGWQNSAMQSRDRRNMTLLGDLVLSNKLVLYDLENQLIGWTEYNCSSSIK 443
Query: 266 VKDEKTGIVHTVGAHKISSSSTYIVGRILTFFLLI 162
VKDE TG VH VG+H ISS+ L F LL+
Sbjct: 444 VKDEGTGTVHLVGSHFISSALPLDTSMCLLFSLLL 478
>ref|NP_198475.1| aspartyl protease-like; protein id: At5g36260.1 [Arabidopsis
thaliana] gi|9759039|dbj|BAB09366.1| aspartyl
protease-like [Arabidopsis thaliana]
Length = 478
Score = 167 bits (423), Expect = 1e-40
Identities = 82/161 (50%), Positives = 116/161 (71%), Gaps = 1/161 (0%)
Frame = -3
Query: 623 VYDQLMSKVLAKQPRLKVYLVEEQYSCFQYTGNVDSGFPIVKLHFEDSLSLTVYPHDYLF 444
+Y+ L+ K+ AKQ ++K+++V+E ++CF +T N D FP+V LHFEDSL L+VYPHDYLF
Sbjct: 318 LYNSLIEKITAKQ-QVKLHMVQETFACFSFTSNTDKAFPVVNLHFEDSLKLSVYPHDYLF 376
Query: 443 NYKGDSYWCIGWQKSASETKNGKDMTLLGDFVLSNKLVVYDLENMTIGWTDYNCSSSIKV 264
+ + D Y C GWQ T++G D+ LLGD VLSNKLVVYDLEN IGW D+NCSSSIKV
Sbjct: 377 SLREDMY-CFGWQSGGMTTQDGADVILLGDLVLSNKLVVYDLENEVIGWADHNCSSSIKV 435
Query: 263 KDEKTGIVHTVGAHK-ISSSSTYIVGRILTFFLLISAMLNS 144
KD +G + +GA IS++S+ + G ++T ++ + +S
Sbjct: 436 KD-GSGAAYQLGAENLISAASSVMNGTLVTLLSILIWVFHS 475
>gb|AAM20468.1| putative aspartyl protease [Arabidopsis thaliana]
gi|23198124|gb|AAN15589.1| putative aspartyl protease
[Arabidopsis thaliana]
Length = 320
Score = 166 bits (421), Expect = 2e-40
Identities = 75/154 (48%), Positives = 108/154 (69%)
Frame = -3
Query: 623 VYDQLMSKVLAKQPRLKVYLVEEQYSCFQYTGNVDSGFPIVKLHFEDSLSLTVYPHDYLF 444
VY+ L++++LA P L ++ V+E ++CF YT +D FP V F+ S+SL VYP +YLF
Sbjct: 160 VYNPLLNEILASHPELTLHTVQESFTCFHYTDKLDR-FPTVTFQFDKSVSLAVYPREYLF 218
Query: 443 NYKGDSYWCIGWQKSASETKNGKDMTLLGDFVLSNKLVVYDLENMTIGWTDYNCSSSIKV 264
+ D+ WC GWQ +TK G +T+LGD LSNKLVVYD+EN IGWT++NCS I+V
Sbjct: 219 QVREDT-WCFGWQNGGLQTKGGASLTILGDMALSNKLVVYDIENQVIGWTNHNCSGGIQV 277
Query: 263 KDEKTGIVHTVGAHKISSSSTYIVGRILTFFLLI 162
KDE++G ++TVGAH +S SS+ + ++LT L+
Sbjct: 278 KDEESGAIYTVGAHNLSWSSSLAITKLLTLVSLL 311
>ref|NP_186923.1| putative aspartyl protease; protein id: At3g02740.1, supported by
cDNA: 40409. [Arabidopsis thaliana]
gi|6728988|gb|AAF26986.1|AC018363_31 putative aspartyl
protease [Arabidopsis thaliana]
gi|21593593|gb|AAM65560.1| putative aspartyl protease
[Arabidopsis thaliana]
Length = 488
Score = 166 bits (421), Expect = 2e-40
Identities = 75/154 (48%), Positives = 108/154 (69%)
Frame = -3
Query: 623 VYDQLMSKVLAKQPRLKVYLVEEQYSCFQYTGNVDSGFPIVKLHFEDSLSLTVYPHDYLF 444
VY+ L++++LA P L ++ V+E ++CF YT +D FP V F+ S+SL VYP +YLF
Sbjct: 328 VYNPLLNEILASHPELTLHTVQESFTCFHYTDKLDR-FPTVTFQFDKSVSLAVYPREYLF 386
Query: 443 NYKGDSYWCIGWQKSASETKNGKDMTLLGDFVLSNKLVVYDLENMTIGWTDYNCSSSIKV 264
+ D+ WC GWQ +TK G +T+LGD LSNKLVVYD+EN IGWT++NCS I+V
Sbjct: 387 QVREDT-WCFGWQNGGLQTKGGASLTILGDMALSNKLVVYDIENQVIGWTNHNCSGGIQV 445
Query: 263 KDEKTGIVHTVGAHKISSSSTYIVGRILTFFLLI 162
KDE++G ++TVGAH +S SS+ + ++LT L+
Sbjct: 446 KDEESGAIYTVGAHNLSWSSSLAITKLLTLVSLL 479
>ref|NP_176703.1| hypothetical protein; protein id: At1g65240.1 [Arabidopsis
thaliana]
Length = 475
Score = 155 bits (393), Expect = 4e-37
Identities = 77/152 (50%), Positives = 106/152 (69%), Gaps = 1/152 (0%)
Frame = -3
Query: 629 RIVYDQLMSKVLAKQPRLKVYLVEEQYSCFQYTGNVDSGFPIVKLHFEDSLSLTVYPHDY 450
+++YD L+ +LA+QP +K+++VEE + CF ++ NVD FP V FEDS+ LTVYPHDY
Sbjct: 313 KVLYDSLIETILARQP-VKLHIVEETFQCFSFSTNVDEAFPPVSFEFEDSVKLTVYPHDY 371
Query: 449 LFNYKGDSYWCIGWQKSASETKNGKDMTLLGDFVLSNKLVVYDLENMTIGWTDYNCSSSI 270
LF + + Y C GWQ T ++ LLGD VLSNKLVVYDL+N IGW D+NCSSSI
Sbjct: 372 LFTLEEELY-CFGWQAGGLTTDERSEVILLGDLVLSNKLVVYDLDNEVIGWADHNCSSSI 430
Query: 269 KVKDEKTGIVHTVGAHKISSSSTYI-VGRILT 177
K+KD G V++VGA +SS+ + + ++LT
Sbjct: 431 KIKDGSGG-VYSVGADNLSSAPRLLMITKLLT 461
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 569,996,784
Number of Sequences: 1393205
Number of extensions: 12081852
Number of successful extensions: 27194
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 26364
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27159
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)