Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004306A_C01 KMC004306A_c01
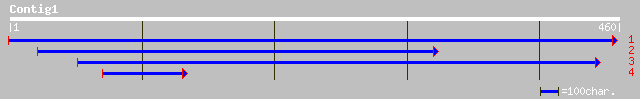
(459 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN84503.1| serine/threonine protein kinase [Oryza sativa (ja... 104 4e-22
ref|NP_194846.1| protein kinase - like protein; protein id: At4g... 99 3e-20
ref|NP_565568.1| putative protein kinase; protein id: At2g24360.... 96 2e-19
pir||G84635 probable protein kinase [imported] - Arabidopsis tha... 96 2e-19
gb|AAK11734.1| serine/threonine/tyrosine kinase [Arachis hypogaea] 95 4e-19
>gb|AAN84503.1| serine/threonine protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 422
Score = 104 bits (260), Expect = 4e-22
Identities = 45/57 (78%), Positives = 51/57 (88%)
Frame = -3
Query: 457 CLPVLREIMTRCWDPNPDVRPPFAEIVEMLENAQTEIMMTVRKARFRCCMTQPMTTD 287
CLPVL EIMTRCWDPNPDVRPPF E+V MLE+A+ I+ TVRKARFRCC++QPMTTD
Sbjct: 366 CLPVLSEIMTRCWDPNPDVRPPFTEVVRMLEHAEVVILSTVRKARFRCCISQPMTTD 422
>ref|NP_194846.1| protein kinase - like protein; protein id: At4g31170.1, supported
by cDNA: 156374. [Arabidopsis thaliana]
gi|7488261|pir||T10671 protein kinase homolog F6E21.90 -
Arabidopsis thaliana gi|7270019|emb|CAB79835.1| protein
kinase-like protein [Arabidopsis thaliana]
gi|21553666|gb|AAM62759.1| protein kinase-like protein
[Arabidopsis thaliana] gi|21928155|gb|AAM78105.1|
AT4g31170/F6E21_90 [Arabidopsis thaliana]
gi|23308373|gb|AAN18156.1| At4g31170/F6E21_90
[Arabidopsis thaliana]
Length = 412
Score = 98.6 bits (244), Expect = 3e-20
Identities = 45/57 (78%), Positives = 49/57 (85%)
Frame = -3
Query: 457 CLPVLREIMTRCWDPNPDVRPPFAEIVEMLENAQTEIMMTVRKARFRCCMTQPMTTD 287
CLPVL EIMTRCWD +P+VRP FAEIV +LE A+TEIM VRKARFRCCMTQPMT D
Sbjct: 356 CLPVLGEIMTRCWDADPEVRPCFAEIVNLLEAAETEIMTNVRKARFRCCMTQPMTVD 412
>ref|NP_565568.1| putative protein kinase; protein id: At2g24360.1, supported by
cDNA: gi_15028152 [Arabidopsis thaliana]
gi|15028153|gb|AAK76700.1| putative protein kinase
[Arabidopsis thaliana] gi|20197761|gb|AAD18109.2|
putative protein kinase [Arabidopsis thaliana]
gi|22136932|gb|AAM91810.1| putative protein kinase
[Arabidopsis thaliana]
Length = 411
Score = 95.5 bits (236), Expect = 2e-19
Identities = 41/57 (71%), Positives = 49/57 (85%)
Frame = -3
Query: 457 CLPVLREIMTRCWDPNPDVRPPFAEIVEMLENAQTEIMMTVRKARFRCCMTQPMTTD 287
CLPVL +IMTRCWD NP+VRP F E+V++LE A+TEIM T RKARFRCC++QPMT D
Sbjct: 355 CLPVLSDIMTRCWDANPEVRPCFVEVVKLLEAAETEIMTTARKARFRCCLSQPMTID 411
>pir||G84635 probable protein kinase [imported] - Arabidopsis thaliana
Length = 407
Score = 95.5 bits (236), Expect = 2e-19
Identities = 41/57 (71%), Positives = 49/57 (85%)
Frame = -3
Query: 457 CLPVLREIMTRCWDPNPDVRPPFAEIVEMLENAQTEIMMTVRKARFRCCMTQPMTTD 287
CLPVL +IMTRCWD NP+VRP F E+V++LE A+TEIM T RKARFRCC++QPMT D
Sbjct: 351 CLPVLSDIMTRCWDANPEVRPCFVEVVKLLEAAETEIMTTARKARFRCCLSQPMTID 407
>gb|AAK11734.1| serine/threonine/tyrosine kinase [Arachis hypogaea]
Length = 411
Score = 94.7 bits (234), Expect = 4e-19
Identities = 44/57 (77%), Positives = 46/57 (80%)
Frame = -3
Query: 457 CLPVLREIMTRCWDPNPDVRPPFAEIVEMLENAQTEIMMTVRKARFRCCMTQPMTTD 287
CLPVLR+IM RCWDPNPDVRPP I NA+TEIM TVRKARFRCCMTQPMT D
Sbjct: 358 CLPVLRDIMPRCWDPNPDVRPP---ICRNCRNAETEIMTTVRKARFRCCMTQPMTAD 411
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 398,215,941
Number of Sequences: 1393205
Number of extensions: 8395651
Number of successful extensions: 22661
Number of sequences better than 10.0: 709
Number of HSP's better than 10.0 without gapping: 22108
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22658
length of database: 448,689,247
effective HSP length: 112
effective length of database: 292,650,287
effective search space used: 11706011480
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)