Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004305A_C01 KMC004305A_c01
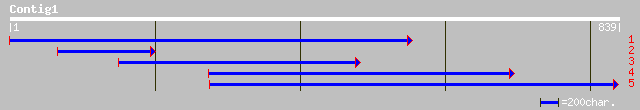
(837 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T52337 phosphoprotein phosphatase (EC 3.1.3.16) 2C [importe... 229 5e-59
ref|NP_175238.1| protein phosphatase-2C, putative; protein id: A... 204 2e-51
dbj|BAB12036.1| putative protein phosphatase [Oryza sativa (japo... 201 8e-51
ref|NP_191785.1| protein phosphatase 2C (PP2C); protein id: At3g... 196 4e-49
dbj|BAB02728.1| protein phosphatase 2C-like protein [Arabidopsis... 192 4e-48
>pir||T52337 phosphoprotein phosphatase (EC 3.1.3.16) 2C [imported] - common ice
plant gi|3643085|gb|AAC36698.1| protein phosphatase-2C;
PP2C [Mesembryanthemum crystallinum]
Length = 359
Score = 229 bits (583), Expect = 5e-59
Identities = 114/170 (67%), Positives = 140/170 (82%)
Frame = -2
Query: 836 KAVDMSHDHRPSYLPERRRVEELGGFIDDGYLNGYLSVTRALGDWDFKLPIGAASPLIAE 657
+A+DMS DHRP+Y E+RRVEELGG++DDGYLNG LSV+RALGDWD KLP G+ASPLI+E
Sbjct: 189 EAIDMSQDHRPTYPSEKRRVEELGGYVDDGYLNGVLSVSRALGDWDMKLPKGSASPLISE 248
Query: 656 PDVQLITLTEDDEFLIIGCDGIWDVKYSQVSPVSLVRRGLRRHDDPQQCARELVKEALRL 477
P+++ I LTEDDEFLIIGCDGIWDV SQ VS+VR GL+RHDDP+Q A++LV EALR
Sbjct: 249 PELRQIILTEDDEFLIIGCDGIWDVISSQ-QAVSIVRWGLKRHDDPEQSAKDLVNEALRR 307
Query: 476 NTSDNLTVIVICLSSIESTVESCPPQRRRFKACSLSEEARNRLKSLMEGN 327
+T DNLTVI++C SSI+ E P+ RRF+ CSLS EA + L+SL+EGN
Sbjct: 308 HTIDNLTVIIVCFSSIDHQREQTGPRPRRFR-CSLSAEALSSLRSLLEGN 356
>ref|NP_175238.1| protein phosphatase-2C, putative; protein id: At1g48040.1
[Arabidopsis thaliana]
gi|8778520|gb|AAF79528.1|AC023673_16 F21D18.27
[Arabidopsis thaliana]
gi|12323084|gb|AAG51521.1|AC051631_1 protein
phosphatase-2C, putative; 42154-43770 [Arabidopsis
thaliana]
Length = 377
Score = 204 bits (518), Expect = 2e-51
Identities = 104/169 (61%), Positives = 134/169 (78%), Gaps = 1/169 (0%)
Frame = -2
Query: 833 AVDMSHDHRPSYLPERRRVEELGGFIDDGYLNGYLSVTRALGDWDFKLPI-GAASPLIAE 657
AVDMS DHR +Y PERRR+E+LGG+ +DGYLNG L+VTRA+GDW+ K P ++SPLI++
Sbjct: 212 AVDMSFDHRSTYEPERRRIEDLGGYFEDGYLNGVLAVTRAIGDWELKNPFTDSSSPLISD 271
Query: 656 PDVQLITLTEDDEFLIIGCDGIWDVKYSQVSPVSLVRRGLRRHDDPQQCARELVKEALRL 477
P++ I LTEDDEFLI+ CDGIWDV SQ + VS VR+GLRRH DP+QCA EL KEA RL
Sbjct: 272 PEIGQIILTEDDEFLILACDGIWDVLSSQ-NAVSNVRQGLRRHGDPRQCAMELGKEAARL 330
Query: 476 NTSDNLTVIVICLSSIESTVESCPPQRRRFKACSLSEEARNRLKSLMEG 330
+SDN+TVIVIC SS+ S+ + PQRRR + C +S+EAR RL++++ G
Sbjct: 331 QSSDNMTVIVICFSSVPSSPKQ--PQRRRLRFC-VSDEARARLQAMLAG 376
>dbj|BAB12036.1| putative protein phosphatase [Oryza sativa (japonica
cultivar-group)]
Length = 380
Score = 201 bits (512), Expect = 8e-51
Identities = 109/174 (62%), Positives = 125/174 (71%), Gaps = 5/174 (2%)
Frame = -2
Query: 833 AVDMSHDHRPSYLPERRRVEELGGFIDDGYLNGYLSVTRALGDWDFKLPIGAASPLIAEP 654
AV+MS DHRP+Y E R+ E GG+I+DGYLNG LSVTRALGDWD K+P G+ SPLIAEP
Sbjct: 209 AVEMSRDHRPTYDAEHERITECGGYIEDGYLNGVLSVTRALGDWDMKMPQGSRSPLIAEP 268
Query: 653 DVQLITLTEDDEFLIIGCDGIWDVKYSQVSPVSLVRRGLRRHDDPQQCARELVKEALRLN 474
+ Q TLTEDDEFLIIGCDGIWDV SQ V++VR+GLRRHDDP++CAREL EA RL
Sbjct: 269 EFQQTTLTEDDEFLIIGCDGIWDVMSSQ-HAVTIVRKGLRRHDDPERCARELAMEAKRLQ 327
Query: 473 TSDNLTVIVICLSSI----ESTVESCPPQRRRFKAC-SLSEEARNRLKSLMEGN 327
T DNLTVIVIC S + E P RR + C SLS EA LK +E N
Sbjct: 328 TFDNLTVIVICFGSELGGGSPSSEQAP--IRRVRCCKSLSSEALCNLKKWLEPN 379
>ref|NP_191785.1| protein phosphatase 2C (PP2C); protein id: At3g62260.1, supported
by cDNA: 20050. [Arabidopsis thaliana]
gi|11358069|pir||T48018 hypothetical protein T17J13.220
- Arabidopsis thaliana gi|6899936|emb|CAB71886.1|
putative protein [Arabidopsis thaliana]
gi|21554078|gb|AAM63159.1| protein phosphatase-2C
[Arabidopsis thaliana]
Length = 383
Score = 196 bits (498), Expect = 4e-49
Identities = 102/170 (60%), Positives = 129/170 (75%), Gaps = 1/170 (0%)
Frame = -2
Query: 836 KAVDMSHDHRPSYLPERRRVEELGGFI-DDGYLNGYLSVTRALGDWDFKLPIGAASPLIA 660
+A+DMS DH+P L ERRRVEE GGFI +DGYLN L+VTRALGDWD KLP G+ SPLI+
Sbjct: 216 RAIDMSEDHKPINLLERRRVEESGGFITNDGYLNEVLAVTRALGDWDLKLPHGSQSPLIS 275
Query: 659 EPDVQLITLTEDDEFLIIGCDGIWDVKYSQVSPVSLVRRGLRRHDDPQQCARELVKEALR 480
EP+++ ITLTEDDEFL+IGCDGIWDV SQ VS+VRRGL RH+DP +CARELV EAL
Sbjct: 276 EPEIKQITLTEDDEFLVIGCDGIWDVLTSQ-EAVSIVRRGLNRHNDPTRCARELVMEALG 334
Query: 479 LNTSDNLTVIVICLSSIESTVESCPPQRRRFKACSLSEEARNRLKSLMEG 330
N+ DNLT +V+C +++ + P +R + SLS EA L++L++G
Sbjct: 335 RNSFDNLTAVVVCFMTMDRGDKPVVPLEKR-RCFSLSPEAFCSLRNLLDG 383
>dbj|BAB02728.1| protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 422
Score = 192 bits (489), Expect = 4e-48
Identities = 101/172 (58%), Positives = 134/172 (77%), Gaps = 3/172 (1%)
Frame = -2
Query: 836 KAVDMSHDHRPSYLPERRRVEELGGFIDDGYLNGYLSVTRALGDWDFK--LPIG-AASPL 666
KAVDMS DH+ ++ PERRRVE+LGG+ + YL G L+VTRALGDW K P+G + SPL
Sbjct: 259 KAVDMSFDHKSTFEPERRRVEDLGGYFEGEYLYGDLAVTRALGDWSIKRFSPLGESLSPL 318
Query: 665 IAEPDVQLITLTEDDEFLIIGCDGIWDVKYSQVSPVSLVRRGLRRHDDPQQCARELVKEA 486
I++PD+Q + LTE+DEFLI+GCDG+WDV SQ + V+ VR+GLRRH DP++CA EL +EA
Sbjct: 319 ISDPDIQQMILTEEDEFLIMGCDGVWDVMTSQYA-VTFVRQGLRRHGDPRRCAMELGREA 377
Query: 485 LRLNTSDNLTVIVICLSSIESTVESCPPQRRRFKACSLSEEARNRLKSLMEG 330
LRL++SDN+TV+VIC SS S PQRRR + C +S+EAR RL++++EG
Sbjct: 378 LRLDSSDNVTVVVICFSS------SPAPQRRRIRFC-VSDEARARLQTMLEG 422
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 718,923,316
Number of Sequences: 1393205
Number of extensions: 15353751
Number of successful extensions: 38801
Number of sequences better than 10.0: 438
Number of HSP's better than 10.0 without gapping: 37084
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38415
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 43480044972
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)